A stromal cell niche sustains ILC2-mediated type-2 conditioning in adipose tissue
- PMID: 31248899
- PMCID: PMC6719433
- DOI: 10.1084/jem.20190689
A stromal cell niche sustains ILC2-mediated type-2 conditioning in adipose tissue
Abstract
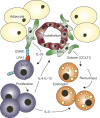
Group-2 innate lymphoid cells (ILC2), type-2 cytokines, and eosinophils have all been implicated in sustaining adipose tissue homeostasis. However, the interplay between the stroma and adipose-resident immune cells is less well understood. We identify that white adipose tissue-resident multipotent stromal cells (WAT-MSCs) can act as a reservoir for IL-33, especially after cell stress, but also provide additional signals for sustaining ILC2. Indeed, we demonstrate that WAT-MSCs also support ICAM-1-mediated proliferation and activation of LFA-1-expressing ILC2s. Consequently, ILC2-derived IL-4 and IL-13 feed back to induce eotaxin secretion from WAT-MSCs, supporting eosinophil recruitment. Thus, MSCs provide a niche for multifaceted dialogue with ILC2 to sustain a type-2 immune environment in WAT.
© 2019 Rana et al.
Figures




Comment in
-
ILC2s chew the fat.J Exp Med. 2019 Sep 2;216(9):1972-1973. doi: 10.1084/jem.20191098. Epub 2019 Aug 12. J Exp Med. 2019. PMID: 31405894 Free PMC article.
References
-
- Berton G., Laudanna C., Sorio C., and Rossi F.. 1992. Generation of signals activating neutrophil functions by leukocyte integrins: LFA-1 and gp150/95, but not CR3, are able to stimulate the respiratory burst of human neutrophils. J. Cell Biol. 116:1007–1017. 10.1083/jcb.116.4.1007 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_12012/2/MRC_/Medical Research Council/United Kingdom
- MC_UU_00014/5/MRC_/Medical Research Council/United Kingdom
- MC_UU_00014/2/MRC_/Medical Research Council/United Kingdom
- MC_U105178805/MRC_/Medical Research Council/United Kingdom
- G0600717/MRC_/Medical Research Council/United Kingdom
- U105178805/MRC_/Medical Research Council/United Kingdom
- G0802051/MRC_/Medical Research Council/United Kingdom
- MC_UU_12012/5/MRC_/Medical Research Council/United Kingdom
- G0400192/MRC_/Medical Research Council/United Kingdom
- RG/12/13/29853/BHF_/British Heart Foundation/United Kingdom
- RG/18/7/33636/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

