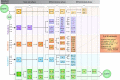
A periodic table of cell types
- PMID: 31249003
- PMCID: PMC6602355
- DOI: 10.1242/dev.169854
A periodic table of cell types
Abstract
Single cell biology is currently revolutionizing developmental and evolutionary biology, revealing new cell types and states in an impressive range of biological systems. With the accumulation of data, however, the field is grappling with a central unanswered question: what exactly is a cell type? This question is further complicated by the inherently dynamic nature of developmental processes. In this Hypothesis article, we propose that a 'periodic table of cell types' can be used as a framework for distinguishing cell types from cell states, in which the periods and groups correspond to developmental trajectories and stages along differentiation, respectively. The different states of the same cell type are further analogous to 'isotopes'. We also highlight how the concept of a periodic table of cell types could be useful for predicting new cell types and states, and for recognizing relationships between cell types throughout development and evolution.
Keywords: Cell atlas; Cell states; Cell types; Periodic table; Single cell RNA-seq.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
-
- Alberts B., Johnson A., Lewis J., Morgan D., Raff M., Roberts K. and Walter P. (2014). The innate and adaptive immune systems. In Molecular Biology of the Cell, 6th edn, pp. 1297-1342. Garland Science.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

