Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features
- PMID: 31249028
- PMCID: PMC6731024
- DOI: 10.1126/science.aaw2872
Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features
Abstract
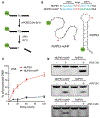
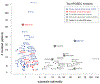
Cancer drivers require statistical modeling to distinguish them from passenger events, which accumulate during tumorigenesis but provide no fitness advantage to cancer cells. The discovery of driver genes and mutations relies on the assumption that exact positional recurrence is unlikely by chance; thus, the precise sharing of mutations across patients identifies drivers. Examining the mutation landscape in cancer genomes, we found that many recurrent cancer mutations previously designated as drivers are likely passengers. Our integrated bioinformatic and biochemical analyses revealed that these passenger hotspot mutations arise from the preference of APOBEC3A, a cytidine deaminase, for DNA stem-loops. Conversely, recurrent APOBEC-signature mutations not in stem-loops are enriched in well-characterized driver genes and may predict new drivers. This demonstrates that mesoscale genomic features need to be integrated into computational models aimed at identifying mutations linked to diseases.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


Comment in
-
Mutation hotspots may not be drug targets.Science. 2019 Jun 28;364(6447):1228-1229. doi: 10.1126/science.aax9108. Science. 2019. PMID: 31249043 No abstract available.
-
Passengers masquerading as cancer drivers.Nat Rev Cancer. 2019 Sep;19(9):485. doi: 10.1038/s41568-019-0184-y. Nat Rev Cancer. 2019. PMID: 31337868 No abstract available.

