Multi-omics Biomarker Pipeline Reveals Elevated Levels of Protein-glutamine Gamma-glutamyltransferase 4 in Seminal Plasma of Prostate Cancer Patients
- PMID: 31249104
- PMCID: PMC6731075
- DOI: 10.1074/mcp.RA119.001612
Multi-omics Biomarker Pipeline Reveals Elevated Levels of Protein-glutamine Gamma-glutamyltransferase 4 in Seminal Plasma of Prostate Cancer Patients
Abstract
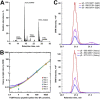
Seminal plasma, because of its proximity to prostate, is a promising fluid for biomarker discovery and noninvasive diagnostics. In this study, we investigated if seminal plasma proteins could increase diagnostic specificity of detecting primary prostate cancer and discriminate between high- and low-grade cancers. To select 147 most promising biomarker candidates, we combined proteins identified through five independent experimental or data mining approaches: tissue transcriptomics, seminal plasma proteomics, cell line secretomics, tissue specificity, and androgen regulation. A rigorous biomarker development pipeline based on selected reaction monitoring assays was designed to evaluate the most promising candidates. As a result, we qualified 76, and verified 19 proteins in seminal plasma of 67 negative biopsy and 152 prostate cancer patients. Verification revealed a prostate-specific, secreted and androgen-regulated protein-glutamine gamma-glutamyltransferase 4 (TGM4), which predicted prostate cancer on biopsy and outperformed age and serum Prostate-Specific Antigen (PSA). A machine-learning approach for data analysis provided improved multi-marker combinations for diagnosis and prognosis. In the independent verification set measured by an in-house immunoassay, TGM4 protein was upregulated 3.7-fold (p = 0.006) and revealed AUC = 0.66 for detecting prostate cancer on biopsy for patients with serum PSA ≥4 ng/ml and age ≥50. Very low levels of TGM4 (120 pg/ml) were detected in blood serum. Collectively, our study demonstrated rigorous evaluation of one of the remaining and not well-explored prostate-specific proteins within the medium-abundance proteome of seminal plasma. Performance of TGM4 warrants its further investigation within the distinct genomic subtypes and evaluation for the inclusion into emerging multi-biomarker panels.
Keywords: Prostate cancer biomarkers; TGM4; XGBoost; absolute quantification; assay development; machine learning; protein-glutamine gamma-glutamyltransferase 4; selected reaction monitoring; seminal plasma; serum/plasma.
© 2019 Drabovich et al.
Figures




Similar articles
-
Seminal fluid: a useful source of prostate cancer biomarkers?Biomark Med. 2015;9(2):77-80. doi: 10.2217/bmm.14.110. Biomark Med. 2015. PMID: 25689895 No abstract available.
-
Seminal plasma as a source of prostate cancer peptide biomarker candidates for detection of indolent and advanced disease.PLoS One. 2013 Jun 24;8(6):e67514. doi: 10.1371/journal.pone.0067514. Print 2013. PLoS One. 2013. PMID: 23826311 Free PMC article.
-
Zinc α2-glycoprotein as a potential novel urine biomarker for the early diagnosis of prostate cancer.BJU Int. 2012 Dec;110(11 Pt B):E688-93. doi: 10.1111/j.1464-410X.2012.11501.x. Epub 2012 Sep 28. BJU Int. 2012. PMID: 23020913
-
Seminal plasma as a diagnostic fluid for male reproductive system disorders.Nat Rev Urol. 2014 May;11(5):278-88. doi: 10.1038/nrurol.2014.74. Epub 2014 Apr 8. Nat Rev Urol. 2014. PMID: 24709963 Review.
-
Semen as a rich source of diagnostic biomarkers for prostate cancer: latest evidence and implications.Mol Cell Biochem. 2022 Jan;477(1):213-223. doi: 10.1007/s11010-021-04273-4. Epub 2021 Oct 16. Mol Cell Biochem. 2022. PMID: 34655417 Review.
Cited by
-
Prostate cancer in omics era.Cancer Cell Int. 2022 Sep 5;22(1):274. doi: 10.1186/s12935-022-02691-y. Cancer Cell Int. 2022. PMID: 36064406 Free PMC article.
-
Rational Design and Development of SARS-CoV-2 Serological Diagnostics by Immunoprecipitation-Targeted Proteomics.Anal Chem. 2022 Sep 27;94(38):12990-12999. doi: 10.1021/acs.analchem.2c01325. Epub 2022 Sep 12. Anal Chem. 2022. PMID: 36095284 Free PMC article.
-
Extracellular Vesicle Proteome in Prostate Cancer: A Comparative Analysis of Mass Spectrometry Studies.Int J Mol Sci. 2021 Dec 19;22(24):13605. doi: 10.3390/ijms222413605. Int J Mol Sci. 2021. PMID: 34948404 Free PMC article. Review.
-
Development of a predictive model to distinguish prostate cancer from benign prostatic hyperplasia by integrating serum glycoproteomics and clinical variables.Clin Proteomics. 2023 Nov 21;20(1):52. doi: 10.1186/s12014-023-09439-4. Clin Proteomics. 2023. PMID: 37990292 Free PMC article.
-
Seminal Plasma Proteome as an Indicator of Sperm Dysfunction and Low Sperm Motility in Chickens.Mol Cell Proteomics. 2020 Jun;19(6):1035-1046. doi: 10.1074/mcp.RA120.002017. Epub 2020 Apr 20. Mol Cell Proteomics. 2020. PMID: 32312844 Free PMC article.
References
-
- Jemal A., Bray F., Center M. M., Ferlay J., Ward E., and Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90 - PubMed
-
- Wilt T. J., Brawer M. K., Jones K. M., Barry M. J., Aronson W. J., Fox S, Gingrich J. R., Wei J. T., Gilhooly P., Grob B. M., Nsouli I., Iyer P., Cartagena R., Snider G., Roehrborn C., Sharifi R., Blank W., Pandya P., Andriole G. L., Culkin D., and Wheeler T. (2012) Radical prostatectomy versus observation for localized prostate cancer. N. Engl. J. Med. 367, 203–213 - PMC - PubMed
-
- Schroder F. H., Hugosson J., Roobol M. J., Tammela T. L., Ciatto S., Nelen V., Kwiatkowski M., Lujan M., Lilja H., Zappa M., Denis L. J., Recker F., Berenguer A., Määttänen L., Bangma C. H., Aus G., Villers A., Rebillard X., van der Kwast T., Blijenberg B. G., Moss S. M., de Koning H. J., Auvinen A., and ERSPC Investigators (2009) Screening and prostate-cancer mortality in a randomized European study. N. Engl. J. Med. 360, 1320–1328 - PubMed
-
- McNaughton-Collins M. F., and Barry M. J. (2011) One man at a time–resolving the PSA controversy. N. Engl. J. Med. 365, 1951–1953 - PubMed
-
- Konety B. R., Bird V. Y., Deorah S., and Dahmoush L. (2005) Comparison of the incidence of latent prostate cancer detected at autopsy before and after the prostate specific antigen era. J. Urol. 174, 1785–1788; discussion 1788 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

