How sample heterogeneity can obscure the signal of microbial interactions
- PMID: 31249391
- PMCID: PMC6794314
- DOI: 10.1038/s41396-019-0463-3
How sample heterogeneity can obscure the signal of microbial interactions
Abstract
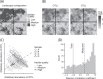
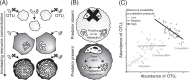
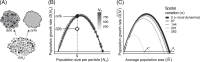
Microbial community data are commonly subjected to computational tools such as correlation networks, null models, and dynamic models, with the goal of identifying the ecological processes structuring microbial communities. A major assumption of these methods is that the signs and magnitudes of species interactions and vital rates can be reliably parsed from observational data on species' (relative) abundances. However, we contend that this assumption is violated when sample units contain any underlying spatial structure. Here, we show how three phenomena-Simpson's paradox, context-dependence, and nonlinear averaging-can lead to erroneous conclusions about population parameters and species interactions when samples contain heterogeneous mixtures of populations or communities. At the root of this issue is the fundamental mismatch between the spatial scales of species interactions (micrometers) and those of typical microbial community samples (millimeters to centimetres). These issues can be overcome by measuring and accounting for spatial heterogeneity at very small scales, which will lead to more reliable inference of the ecological mechanisms structuring natural microbial communities.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures