FCGR3A and FCGR2A Genotypes Differentially Impact Allograft Rejection and Patients' Survival After Lung Transplant
- PMID: 31249568
- PMCID: PMC6582937
- DOI: 10.3389/fimmu.2019.01208
FCGR3A and FCGR2A Genotypes Differentially Impact Allograft Rejection and Patients' Survival After Lung Transplant
Abstract
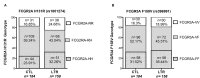
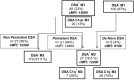
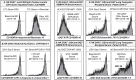
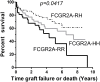
Fc gamma receptors (FcγRs) play a major role in the regulation of humoral immune responses. Single-nucleotide polymorphisms (SNPs) of FCGR2A and FCGR3A can impact the expression level, IgG affinity and function of the CD32 and CD16 FcγRs in response to their engagement by the Fc fragment of IgG. The CD16 isoform encoded by FCGR3A [158V/V] controls the intensity of antibody-dependent cytotoxic alloimmune responses of natural killer cells (NK) and has been identified as a susceptibility marker predisposing patients to cardiac allograft vasculopathy after heart transplant. This study aimed to investigate whether FCGR2A and FCGR3A polymorphisms can also be associated with the clinical outcome of lung transplant recipients (LTRs). The SNPs of FCGR2A ([131R/H], rs1801274) and FCGR3A ([158V/F], rs396991) were identified in 158 LTRs and 184 Controls (CTL). The corresponding distribution of genotypic and allelic combinations was analyzed for potential links with the development of circulating donor-specific anti-HLA alloantibodies (DSA) detected at months 1 and 3 after lung transplant (LTx), the occurrence of acute rejection (AR) and chronic lung allograft dysfunction (CLAD), and the overall survival of LTRs. The FCGR3A [158V/V] genotype was identified as an independent susceptibility factor associated with higher rates of AR during the first trimester after LTx (HR 4.8, p < 0.0001, 95% CI 2.37-9.61), but it could not be associated with the level of CD16- mediated NK cell activation in response to the LTR's DSA, whatever the MFI intensity and C1q binding profiles of the DSA evaluated. The FCGR2A [131R/R] genotype was associated with lower CLAD-free survival of LTRs, independently of the presence of DSA at 3 months (HR 1.8, p = 0.024, 95% CI 1.08-3.03). Our data indicate that FCGR SNPs differentially affect the clinical outcome of LTRs and may be of use to stratify patients at higher risk of experiencing graft rejection. Furthermore, these data suggest that in the LTx setting, specific mechanisms of humoral alloreactivity, which cannot be solely explained by the complement and CD16-mediated pathogenic effects of DSA, may be involved in the development of acute and chronic lung allograft rejection.
Keywords: Fc-gamma receptors; HLA antibodies; allograft rejection; chronic lung allograft dysfunction; lung transplantation; natural killer cells.
Figures

Similar articles
-
Genetic and Functional Profiling of CD16-Dependent Natural Killer Activation Identifies Patients at Higher Risk of Cardiac Allograft Vasculopathy.Circulation. 2018 Mar 6;137(10):1049-1059. doi: 10.1161/CIRCULATIONAHA.117.030435. Epub 2017 Nov 2. Circulation. 2018. PMID: 29097449
-
FCGR2C Q13 and FCGR3A V176 alleles jointly associate with worse natural killer cell-mediated antibody-dependent cellular cytotoxicity and microvascular inflammation in kidney allograft antibody-mediated rejection.Am J Transplant. 2025 Feb;25(2):302-315. doi: 10.1016/j.ajt.2024.09.018. Epub 2024 Sep 26. Am J Transplant. 2025. PMID: 39332679
-
The FCGR3A 158 V/V-genotype is associated with decreased survival of renal allografts with chronic active antibody-mediated rejection.Sci Rep. 2021 Apr 12;11(1):7903. doi: 10.1038/s41598-021-86943-3. Sci Rep. 2021. PMID: 33846428 Free PMC article.
-
The Role of Donor-Specific Antibodies in Intestinal Transplantation: Experience at the University of California Los Angeles and Literature Review.Clin Transpl. 2014:153-9. Clin Transpl. 2014. PMID: 26281140 Review.
-
Association between functional FCGR3A F158V and FCGR2A R131H polymorphisms and responsiveness to rituximab in patients with autoimmune diseases: a meta-analysis.Pharmacogenomics J. 2023 Nov;23(6):210-216. doi: 10.1038/s41397-023-00308-9. Epub 2023 May 6. Pharmacogenomics J. 2023. PMID: 37149714 Review.
Cited by
-
Deep Learning Reveals Key Immunosuppression Genes and Distinct Immunotypes in Periodontitis.Front Genet. 2021 Mar 12;12:648329. doi: 10.3389/fgene.2021.648329. eCollection 2021. Front Genet. 2021. PMID: 33777111 Free PMC article.
-
NKG2D Natural Killer Cell Receptor-A Short Description and Potential Clinical Applications.Cells. 2021 Jun 7;10(6):1420. doi: 10.3390/cells10061420. Cells. 2021. PMID: 34200375 Free PMC article. Review.
-
Correlations of FCGR2A 131R/H and FCGR3A 158V/F Polymorphisms with the Susceptibility of Peri-implantitis in Chinese Han Population.Mol Biotechnol. 2025 Jun;67(6):2254-2261. doi: 10.1007/s12033-024-01193-8. Epub 2024 May 21. Mol Biotechnol. 2025. PMID: 38771420
-
FCGR2A-HH Gene Variants Encoding the Fc Gamma Receptor for the C-Reactive Protein Are Associated with Enhanced Monocyte CD32 Expression and Cardiovascular Events' Recurrence after Primary Acute Coronary Syndrome.Biomedicines. 2022 Feb 19;10(2):495. doi: 10.3390/biomedicines10020495. Biomedicines. 2022. PMID: 35203703 Free PMC article.
-
Functional Fc Gamma Receptor Gene Polymorphisms and Long-Term Kidney Allograft Survival.Front Immunol. 2021 Aug 23;12:724331. doi: 10.3389/fimmu.2021.724331. eCollection 2021. Front Immunol. 2021. PMID: 34497614 Free PMC article.
References
-
- Chambers DC, Yusen RD, Cherikh WS, Goldfarb SB, Kucheryavaya AY, Khusch K, et al. . The registry of the international society for heart and lung transplantation: thirty-fourth adult lung and heart-lung transplantation report-2017; focus theme: allograft ischemic time. J Heart Lung Transplant. (2017) 36:1047–59. 10.1016/j.healun.2017.07.016 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

