Lysine-Specific Histone Demethylase 1A Regulates Macrophage Polarization and Checkpoint Molecules in the Tumor Microenvironment of Triple-Negative Breast Cancer
- PMID: 31249575
- PMCID: PMC6582666
- DOI: 10.3389/fimmu.2019.01351
Lysine-Specific Histone Demethylase 1A Regulates Macrophage Polarization and Checkpoint Molecules in the Tumor Microenvironment of Triple-Negative Breast Cancer
Abstract
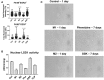
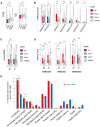
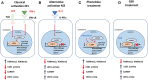
Macrophages play an important role in regulating the tumor microenvironment (TME). Here we show that classical (M1) macrophage polarization reduced expression of LSD1, nuclear REST corepressor 1 (CoREST), and the zinc finger protein SNAIL. The LSD1 inhibitor phenelzine targeted both the flavin adenine dinucleotide (FAD) and CoREST binding domains of LSD1, unlike the LSD1 inhibitor GSK2879552, which only targeted the FAD domain. Phenelzine treatment reduced nuclear demethylase activity and increased transcription and expression of M1-like signatures both in vitro and in a murine triple-negative breast cancer model. Overall, the LSD1 inhibitors phenelzine and GSK2879552 are useful tools for dissecting the contribution of LSD1 demethylase activity and the nuclear LSD1-CoREST complex to switching macrophage polarization programs. These findings suggest that inhibitors must have dual FAD and CoREST targeting abilities to successfully initiate or prime macrophages toward an anti-tumor M1-like phenotype in triple-negative breast cancer.
Keywords: CoREST; LSD1; breast cancer; epigenetics; macrophage polarization; tumor associated macrophages; tumor microenvironment.
Figures





Similar articles
-
Extranucleosomal DNA enhances the activity of the LSD1/CoREST histone demethylase complex.Nucleic Acids Res. 2015 May 26;43(10):4868-80. doi: 10.1093/nar/gkv388. Epub 2015 Apr 27. Nucleic Acids Res. 2015. PMID: 25916846 Free PMC article.
-
Thermodynamic characterization of the binding interaction between the histone demethylase LSD1/KDM1 and CoREST.Biochemistry. 2011 Feb 1;50(4):546-57. doi: 10.1021/bi101776t. Epub 2010 Dec 30. Biochemistry. 2011. PMID: 21142040 Free PMC article.
-
Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate.Mol Cell. 2020 Jun 4;78(5):903-914.e4. doi: 10.1016/j.molcel.2020.04.019. Epub 2020 May 11. Mol Cell. 2020. PMID: 32396821 Free PMC article.
-
Lysine-specific histone demethylase 1 (LSD1): A potential molecular target for tumor therapy.Crit Rev Eukaryot Gene Expr. 2012;22(1):53-9. doi: 10.1615/critreveukargeneexpr.v22.i1.40. Crit Rev Eukaryot Gene Expr. 2012. PMID: 22339659 Review.
-
Lysine-Specific Demethylase 1 (LSD1)-Mediated Epigenetic Modification of Immunogenicity and Immunomodulatory Effects in Breast Cancers.Curr Oncol. 2023 Feb 9;30(2):2127-2143. doi: 10.3390/curroncol30020164. Curr Oncol. 2023. PMID: 36826125 Free PMC article. Review.
Cited by
-
Role of Epigenetic Regulation in Plasticity of Tumor Immune Microenvironment.Front Immunol. 2021 Apr 2;12:640369. doi: 10.3389/fimmu.2021.640369. eCollection 2021. Front Immunol. 2021. PMID: 33868269 Free PMC article. Review.
-
Chemical Language Model Linker: blending text and molecules with modular adapters.ArXiv [Preprint]. 2025 Jun 13:arXiv:2410.20182v3. ArXiv. 2025. Update in: J Chem Inf Model. 2025 Aug 21. doi: 10.1021/acs.jcim.5c00853. PMID: 40321950 Free PMC article. Updated. Preprint.
-
A state-of-the-art review on LSD1 and its inhibitors in breast cancer: Molecular mechanisms and therapeutic significance.Front Pharmacol. 2022 Sep 16;13:989575. doi: 10.3389/fphar.2022.989575. eCollection 2022. Front Pharmacol. 2022. PMID: 36188536 Free PMC article. Review.
-
Transcriptional, Epigenetic and Metabolic Programming of Tumor-Associated Macrophages.Cancers (Basel). 2020 May 29;12(6):1411. doi: 10.3390/cancers12061411. Cancers (Basel). 2020. PMID: 32486098 Free PMC article. Review.
-
Construction and Validation of a Macrophage-Associated Risk Model for Predicting the Prognosis of Osteosarcoma.J Oncol. 2021 Jun 2;2021:9967954. doi: 10.1155/2021/9967954. eCollection 2021. J Oncol. 2021. PMID: 34188683 Free PMC article.
References
-
- Diana A, Franzese E, Centonze S, Carlino F, Della Corte CM, Ventriglia J, et al. . Triple-negative breast cancers: systematic review of the literature on molecular and clinical features with a focus on treatment with innovative drugs. Curr Oncol Rep. (2018) 20:76. 10.1007/s11912-018-0726-6 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

