Genome-Wide Identification and Homoeologous Expression Analysis of PP2C Genes in Wheat (Triticum aestivum L.)
- PMID: 31249596
- PMCID: PMC6582248
- DOI: 10.3389/fgene.2019.00561
Genome-Wide Identification and Homoeologous Expression Analysis of PP2C Genes in Wheat (Triticum aestivum L.)
Abstract
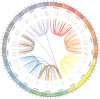
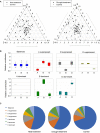
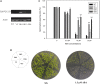
Plant protein phosphatase 2Cs (PP2Cs) play crucial roles in phytohormone signaling, developmental processes, and both biotic and abiotic stress responses. However, little research has been conducted on the PP2C gene family in hexaploid wheat (Triticum aestivum L.), which is an important cereal crop. In this study, a genome-wide investigation of TaPP2C gene family was performed. A total of 257 homoeologs of 95 TaPP2C genes were identified, of which 80% of genes had all the three homoeologs across A, B, and D subgenomes. Domain analysis indicated that all the TaPP2C homoeologs harbored the type 2C phosphatase domains. Based on the phylogenetic analysis, TaPP2Cs were divided into 13 groups (A-M) and 4 single branches, which corresponded to the results of gene structure and protein motif analyses. Results of chromosomal location and synteny relationship analysis of TaPP2C homoeologs revealed that known chromosome translocation events and pericentromeric inversions were responsible for the formation of TaPP2C gene family. Expression patterns of TaPP2C homoeologs in various tissues and under diverse stress conditions were analyzed using publicly available RNA-seq data. The results suggested that TaPP2C genes regulate wheat developmental processes and stress responses. Homoeologous expression patterns of TaPP2C triad homoeologs from A, B, and D subgenomes, revealed expression bias within triads under the normal condition, and variability in expression under different stress treatments. Quantitative real-time PCR (qRT-PCR) analysis of eight TaPP2C genes in group A revealed that they were all up-regulated after abscisic acid treatment. Some genes in group A also responded to other phytohormones such as methyl jasmonate and gibberellin. Yeast two-hybrid assays showed that group A TaPP2Cs also interacted with TaSnRK2.1 and TaSnRK2.2 from subclass II, besides with subclass III TaSnRK2s. TaPP2C135 in group A was transformed into Arabidopsis and germination assay revealed that ectopic expression of TaPP2C135 in Arabidopsis enhanced its tolerance to ABA. Overall, these results enhance our understanding of the function of TaPP2Cs in wheat, and provide novel insights into the roles of group A TaPP2Cs. This information will be useful for in-depth functional analysis of TaPP2Cs in future studies and for wheat breeding.
Keywords: gene expression; genome-wide; homoeologous pattern; protein phosphatase 2C (PP2C); stress response; wheat.
Figures






References
-
- Akimoto-Tomiyama C., Tanabe S., Kajiwara H., Minami E., Ochiai H. (2018). Loss of chloroplast-localized protein phosphatase 2Cs in Arabidopsis thaliana leads to enhancement of plant immunity and resistance to Xanthomonas campestris pv. campestris infection. Mol. Plant Pathol. 19 1184–1195. 10.1111/mpp.12596 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

