De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication
- PMID: 31249862
- PMCID: PMC6594761
- DOI: 10.1126/sciadv.aav0547
De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication
Abstract
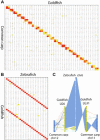
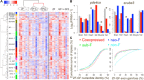
For over a thousand years, the common goldfish (Carassius auratus) was raised throughout Asia for food and as an ornamental pet. As a very close relative of the common carp (Cyprinus carpio), goldfish share the recent genome duplication that occurred approximately 14 million years ago in their common ancestor. The combination of centuries of breeding and a wide array of interesting body morphologies provides an exciting opportunity to link genotype to phenotype and to understand the dynamics of genome evolution and speciation. We generated a high-quality draft sequence and gene annotations of a "Wakin" goldfish using 71X PacBio long reads. The two subgenomes in goldfish retained extensive synteny and collinearity between goldfish and zebrafish. However, genes were lost quickly after the carp whole-genome duplication, and the expression of 30% of the retained duplicated gene diverged substantially across seven tissues sampled. Loss of sequence identity and/or exons determined the divergence of the expression levels across all tissues, while loss of conserved noncoding elements determined expression variance between different tissues. This assembly provides an important resource for comparative genomics and understanding the causes of goldfish variants.
Figures



References
-
- Cerdá-Reverter J. M., Haitina T., Schiöth H. B., Peter R. E., Gene structure of the goldfish agouti-signaling protein: A putative role in the dorsal-ventral pigment pattern of fish. Endocrinology 146, 1597–1610 (2005). - PubMed
-
- Munkittrick K. R., Moccia R. D., Leatherland J. F., Polycystic kidney disease in goldfish (Carassius auratus) from Hamilton Harbour, Lake Ontario, Canada. Vet. Pathol. 22, 232–237 (1985). - PubMed
-
- Sahoo P. K., Swaminathan T. R., Abraham T. J., Kumar R., Pattanayak S., Mohapatra A., Rath S. S., Patra A., Adikesavalu H., Sood N., Pradhan P. K., Das B. K., Jayasankar P., Jena J. K., Detection of goldfish haematopoietic necrosis herpes virus (Cyprinid herpesvirus-2) with multi-drug resistant Aeromonas hydrophila infection in goldfish: First evidence of any viral disease outbreak in ornamental freshwater aquaculture farms in India. Acta Trop. 161, 8–17 (2016). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

