Nutrient availability shapes methionine metabolism in p16/ MTAP-deleted cells
- PMID: 31249865
- PMCID: PMC6594760
- DOI: 10.1126/sciadv.aav7769
Nutrient availability shapes methionine metabolism in p16/ MTAP-deleted cells
Abstract
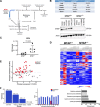
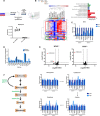
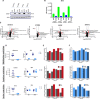
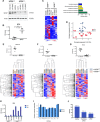
Codeletions of gene loci containing tumor suppressors and neighboring metabolic enzymes present an attractive synthetic dependency in cancers. However, the impact that these genetic events have on metabolic processes, which are also dependent on nutrient availability and other environmental factors, is unknown. As a proof of concept, we considered panels of cancer cells with homozygous codeletions in CDKN2a and MTAP, genes respectively encoding the commonly-deleted tumor suppressor p16 and an enzyme involved in methionine metabolism. A comparative metabolomics analysis revealed that while a metabolic signature of MTAP deletion is apparent, it is not preserved upon restriction of nutrients related to methionine metabolism. Furthermore, re-expression of MTAP exerts heterogeneous consequences on metabolism across isogenic cell pairs. Together, this study demonstrates that numerous factors, particularly nutrition, can overwhelm the effects of metabolic gene deletions on metabolism. These findings may also have relevance to drug development efforts aiming to target methionine metabolism.
Figures


References
-
- Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., Mc Henry K. T., Pinchback R. M., Ligon A. H., Cho Y.-J., Haery L., Greulich H., Reich M., Winckler W., Lawrence M. S., Weir B. A., Tanaka K. E., Chiang D. Y., Bass A. J., Loo A., Hoffman C., Prensner J., Liefeld T., Gao Q., Yecies D., Signoretti S., Maher E., Kaye F. J., Sasaki H., Tepper J. E., Fletcher J. A., Tabernero J., Baselga J., Tsao M.-S., Demichelis F., Rubin M. A., Janne P. A., Daly M. J., Nucera C., Levine R. L., Ebert B. L., Gabriel S., Rustgi A. K., Antonescu C. R., Ladanyi M., Letai A., Garraway L. A., Loda M., Beer D. G., True L. D., Okamoto A., Pomeroy S. L., Singer S., Golub T. R., Lander E. S., Getz G., Sellers W. R., Meyerson M., The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). - PMC - PubMed
-
- Zhang H., Chen Z.-H., Savarese T. M., Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-α1, interferon-β1, and other 9p21 markers in human malignant cell lines. Cancer Genet. Cytogenet. 86, 22–28 (1996). - PubMed
-
- de Oliveira S. F. V., Ganzinelli M., Chilà R., Serino L., Maciel M. E., de Andrade Urban C., de Lima R. S., Cavalli I. J., Generali D., Broggini M., Damia G., de Souza Fonseca Ribeiro E. M., Characterization of MTAP gene expression in breast cancer patients and cell lines. PLOS ONE 11, e0145647 (2016). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

