Enhancer-mediated enrichment of interacting JMJD3-DDX21 to ENPP2 locus prevents R-loop formation and promotes transcription
- PMID: 31251802
- PMCID: PMC6895255
- DOI: 10.1093/nar/gkz560
Enhancer-mediated enrichment of interacting JMJD3-DDX21 to ENPP2 locus prevents R-loop formation and promotes transcription
Abstract
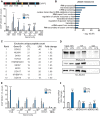
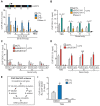
ENPP2, which encodes for the enzyme autotaxin (ATX), is overexpressed during chronic inflammatory diseases and various cancers. However, the molecular mechanism involved in the ENPP2 transcription remains elusive. Here, in HEK 293T cells, we demonstrated that lipopolysaccharide (LPS) increased the transcription process at ENPP2 locus through a NF-кB pathway and a reduction of H3K27me3 level, a histone repressive mark, by the demethylase UTX. Simultaneously, the H3K27me3 demethylase JMJD3/KDM6B was recruited to the transcription start site (TSS), within the gene body and controlled the expression of ENPP2 in a non-enzymatic manner. Mass spectrometry data revealed a novel interaction for JMJD3 with DDX21, a RNA helicase that unwinds R-loops created by nascent transcript and DNA template. Upon LPS treatment, JMJD3 is necessary for DDX21 recruitment at ENPP2 locus allowing the resolution of aberrant R-loops. CRISPR-Cas9-mediated deletion of a distant-acting enhancer decreased the expression of ENPP2 and lowered the recruitment of JMJD3-DDX21 complex at TSS and its progression through the gene body. Taken together, these findings revealed that enhancer-mediated enrichment of novel JMJD3-DDX21 interaction at ENPP2 locus is necessary for nascent transcript synthesis via the resolution of aberrant R-loops formation in response to inflammatory stimulus.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
-
- Barbayianni E., Kaffe E., Aidinis V., Kokotos G.. Autotaxin, a secreted lysophospholipase D, as a promising therapeutic target in chronic inflammation and cancer. Prog. Lipid Res. 2015; 58:76–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

