Symmetric activity of DNA polymerases at and recruitment of exonuclease ExoR and of PolA to the Bacillus subtilis replication forks
- PMID: 31251806
- PMCID: PMC6895272
- DOI: 10.1093/nar/gkz554
Symmetric activity of DNA polymerases at and recruitment of exonuclease ExoR and of PolA to the Bacillus subtilis replication forks
Abstract
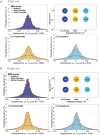
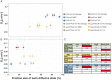
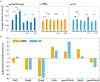
DNA replication forks are intrinsically asymmetric and may arrest during the cell cycle upon encountering modifications in the DNA. We have studied real time dynamics of three DNA polymerases and an exonuclease at a single molecule level in the bacterium Bacillus subtilis. PolC and DnaE work in a symmetric manner and show similar dwell times. After addition of DNA damage, their static fractions and dwell times decreased, in agreement with increased re-establishment of replication forks. Only a minor fraction of replication forks showed a loss of active polymerases, indicating relatively robust activity during DNA repair. Conversely, PolA, homolog of polymerase I and exonuclease ExoR were rarely present at forks during unperturbed replication but were recruited to replications forks after induction of DNA damage. Protein dynamics of PolA or ExoR were altered in the absence of each other during exponential growth and during DNA repair, indicating overlapping functions. Purified ExoR displayed exonuclease activity and preferentially bound to DNA having 5' overhangs in vitro. Our analyses support the idea that two replicative DNA polymerases work together at the lagging strand whilst only PolC acts at the leading strand, and that PolA and ExoR perform inducible functions at replication forks during DNA repair.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






Similar articles
-
Interactions of the Bacillus subtilis DnaE polymerase with replisomal proteins modulate its activity and fidelity.Open Biol. 2017 Sep;7(9):170146. doi: 10.1098/rsob.170146. Open Biol. 2017. PMID: 28878042 Free PMC article.
-
Visual Evidence for the Recruitment of Four Enzymes with RNase Activity to the Bacillus subtilis Replication Forks.Cells. 2024 Aug 20;13(16):1381. doi: 10.3390/cells13161381. Cells. 2024. PMID: 39195267 Free PMC article.
-
The B. subtilis replicative polymerases bind the sliding clamp with different strengths to tune their activity in DNA replication.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf721. doi: 10.1093/nar/gkaf721. Nucleic Acids Res. 2025. PMID: 40737090 Free PMC article.
-
Is there a role for replication fork asymmetry in the distribution of genes in bacterial genomes?Trends Microbiol. 2002 Sep;10(9):393-5. doi: 10.1016/s0966-842x(02)02420-4. Trends Microbiol. 2002. PMID: 12217498 Review.
-
Replicative DNA polymerases.Genome Biol. 2001;2(1):REVIEWS3002. doi: 10.1186/gb-2001-2-1-reviews3002. Epub 2001 Jan 12. Genome Biol. 2001. PMID: 11178285 Free PMC article. Review.
Cited by
-
Reclassification of family A DNA polymerases reveals novel functional subfamilies and distinctive structural features.Nucleic Acids Res. 2023 May 22;51(9):4488-4507. doi: 10.1093/nar/gkad242. Nucleic Acids Res. 2023. PMID: 37070157 Free PMC article.
-
The Replicative DnaE Polymerase of Bacillus subtilis Recruits the Glycolytic Pyruvate Kinase (PykA) When Bound to Primed DNA Templates.Life (Basel). 2023 Apr 7;13(4):965. doi: 10.3390/life13040965. Life (Basel). 2023. PMID: 37109494 Free PMC article.
-
A Bacterial Dynamin-Like Protein Confers a Novel Phage Resistance Strategy on the Population Level in Bacillus subtilis.mBio. 2021 Feb 22;13(1):e0375321. doi: 10.1128/mbio.03753-21. Epub 2022 Feb 15. mBio. 2021. PMID: 35164550 Free PMC article.
-
Single molecule/particle tracking analysis program SMTracker 2.0 reveals different dynamics of proteins within the RNA degradosome complex in Bacillus subtilis.Nucleic Acids Res. 2021 Nov 8;49(19):e112. doi: 10.1093/nar/gkab696. Nucleic Acids Res. 2021. PMID: 34417617 Free PMC article.
-
The Impact of RNA-DNA Hybrids on Genome Integrity in Bacteria.Annu Rev Microbiol. 2022 Sep 8;76:461-480. doi: 10.1146/annurev-micro-102521-014450. Epub 2022 Jun 2. Annu Rev Microbiol. 2022. PMID: 35655343 Free PMC article. Review.
References
-
- Sanders G.M., Dallmann H.G., McHenry C.S.. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol. Cell. 2010; 37:273–281. - PubMed
-
- Henrikus S.S., van Oijen A.M., Robinson A.. Specialised DNA polymerases in Escherichia coli: roles within multiple pathways. Curr. Genet. 2018; 64:1189–1196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

