FastqCleaner: an interactive Bioconductor application for quality-control, filtering and trimming of FASTQ files
- PMID: 31253077
- PMCID: PMC6599294
- DOI: 10.1186/s12859-019-2961-8
FastqCleaner: an interactive Bioconductor application for quality-control, filtering and trimming of FASTQ files
Abstract
Background: Exploration and processing of FASTQ files are the first steps in state-of-the-art data analysis workflows of Next Generation Sequencing (NGS) platforms. The large amount of data generated by these technologies has put a challenge in terms of rapid analysis and visualization of sequencing information. Recent integration of the R data analysis platform with web visual frameworks has stimulated the development of user-friendly, powerful, and dynamic NGS data analysis applications.
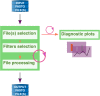
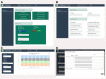
Results: This paper presents FastqCleaner, a Bioconductor visual application for both quality-control (QC) and pre-processing of FASTQ files. The interface shows diagnostic information for the input and output data and allows to select a series of filtering and trimming operations in an interactive framework. FastqCleaner combines the technology of Bioconductor for NGS data analysis with the data visualization advantages of a web environment.
Conclusions: FastqCleaner is an user-friendly, offline-capable tool that enables access to advanced Bioconductor infrastructure. The novel concept of a Bioconductor interactive application that can be used without the need for programming skills, makes FastqCleaner a valuable resource for NGS data analysis.
Keywords: Bioconductor; FASTQ; Next generation sequencing; R; Shiny; User-friendly tool; Visualization; Web app.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
ClinQC: a tool for quality control and cleaning of Sanger and NGS data in clinical research.BMC Bioinformatics. 2016 Feb 2;17:56. doi: 10.1186/s12859-016-0915-y. BMC Bioinformatics. 2016. PMID: 26830926 Free PMC article.
-
systemPipeR: NGS workflow and report generation environment.BMC Bioinformatics. 2016 Sep 20;17:388. doi: 10.1186/s12859-016-1241-0. BMC Bioinformatics. 2016. PMID: 27650223 Free PMC article.
-
GENAVi: a shiny web application for gene expression normalization, analysis and visualization.BMC Genomics. 2019 Oct 16;20(1):745. doi: 10.1186/s12864-019-6073-7. BMC Genomics. 2019. PMID: 31619158 Free PMC article.
-
Using R and Bioconductor in Clinical Genomics and Transcriptomics.J Mol Diagn. 2020 Jan;22(1):3-20. doi: 10.1016/j.jmoldx.2019.08.006. Epub 2019 Oct 9. J Mol Diagn. 2020. PMID: 31605800 Review.
-
Visual programming for next-generation sequencing data analytics.BioData Min. 2016 Apr 27;9:16. doi: 10.1186/s13040-016-0095-3. eCollection 2016. BioData Min. 2016. PMID: 27127540 Free PMC article. Review.
Cited by
-
Comparative Analysis and Identification of Terpene Synthase Genes in Convallaria keiskei Leaf, Flower and Root Using RNA-Sequencing Profiling.Plants (Basel). 2023 Jul 28;12(15):2797. doi: 10.3390/plants12152797. Plants (Basel). 2023. PMID: 37570951 Free PMC article.
-
LABRADOR-A Computational Workflow for Virus Detection in High-Throughput Sequencing Data.Viruses. 2021 Dec 18;13(12):2541. doi: 10.3390/v13122541. Viruses. 2021. PMID: 34960810 Free PMC article.
-
Endogenous Retrovirus Elements Are Co-Expressed with IFN Stimulation Genes in the JAK-STAT Pathway.Viruses. 2022 Dec 24;15(1):60. doi: 10.3390/v15010060. Viruses. 2022. PMID: 36680099 Free PMC article.
-
Microbiota of pest insect Nezara viridula mediate detoxification and plant defense repression.ISME J. 2024 Jan 8;18(1):wrae097. doi: 10.1093/ismejo/wrae097. ISME J. 2024. PMID: 38836495 Free PMC article.
-
Transcriptome-Wide Identification and Quantification of Caffeoylquinic Acid Biosynthesis Pathway and Prediction of Its Putative BAHDs Gene Complex in A. spathulifolius.Int J Mol Sci. 2021 Jun 13;22(12):6333. doi: 10.3390/ijms22126333. Int J Mol Sci. 2021. PMID: 34199260 Free PMC article.
References
-
- Tripathi R, Sharma P, Chakraborty P, Varadwaj P. Next-generation sequencing revolution through big data analytics. Front Life Sci. 2016;9:119–149. doi: 10.1080/21553769.2016.1178180. - DOI
-
- R Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2017.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

