Cell segmentation methods for label-free contrast microscopy: review and comprehensive comparison
- PMID: 31253078
- PMCID: PMC6599268
- DOI: 10.1186/s12859-019-2880-8
Cell segmentation methods for label-free contrast microscopy: review and comprehensive comparison
Abstract
Background: Because of its non-destructive nature, label-free imaging is an important strategy for studying biological processes. However, routine microscopic techniques like phase contrast or DIC suffer from shadow-cast artifacts making automatic segmentation challenging. The aim of this study was to compare the segmentation efficacy of published steps of segmentation work-flow (image reconstruction, foreground segmentation, cell detection (seed-point extraction) and cell (instance) segmentation) on a dataset of the same cells from multiple contrast microscopic modalities.
Results: We built a collection of routines aimed at image segmentation of viable adherent cells grown on the culture dish acquired by phase contrast, differential interference contrast, Hoffman modulation contrast and quantitative phase imaging, and we performed a comprehensive comparison of available segmentation methods applicable for label-free data. We demonstrated that it is crucial to perform the image reconstruction step, enabling the use of segmentation methods originally not applicable on label-free images. Further we compared foreground segmentation methods (thresholding, feature-extraction, level-set, graph-cut, learning-based), seed-point extraction methods (Laplacian of Gaussians, radial symmetry and distance transform, iterative radial voting, maximally stable extremal region and learning-based) and single cell segmentation methods. We validated suitable set of methods for each microscopy modality and published them online.
Conclusions: We demonstrate that image reconstruction step allows the use of segmentation methods not originally intended for label-free imaging. In addition to the comprehensive comparison of methods, raw and reconstructed annotated data and Matlab codes are provided.
Keywords: Cell segmentation; Differential contrast image; Image reconstruction; Laplacian of Gaussians; Methods comparison; Microscopy; Quantitative phase imaging.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



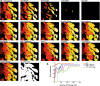
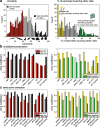
Similar articles
-
Empirical gradient threshold technique for automated segmentation across image modalities and cell lines.J Microsc. 2015 Oct;260(1):86-99. doi: 10.1111/jmi.12269. Epub 2015 Jun 5. J Microsc. 2015. PMID: 26046924
-
LIVECell-A large-scale dataset for label-free live cell segmentation.Nat Methods. 2021 Sep;18(9):1038-1045. doi: 10.1038/s41592-021-01249-6. Epub 2021 Aug 30. Nat Methods. 2021. PMID: 34462594 Free PMC article.
-
Automated method for the rapid and precise estimation of adherent cell culture characteristics from phase contrast microscopy images.Biotechnol Bioeng. 2014 Mar;111(3):504-17. doi: 10.1002/bit.25115. Epub 2013 Oct 5. Biotechnol Bioeng. 2014. PMID: 24037521 Free PMC article.
-
Optimized detection and segmentation of nuclei in gastric cancer images using stain normalization and blurred artifact removal.Pathol Res Pract. 2023 Aug;248:154694. doi: 10.1016/j.prp.2023.154694. Epub 2023 Jul 16. Pathol Res Pract. 2023. PMID: 37494804 Review.
-
Review of research on the instance segmentation of cell images.Comput Methods Programs Biomed. 2022 Dec;227:107211. doi: 10.1016/j.cmpb.2022.107211. Epub 2022 Oct 31. Comput Methods Programs Biomed. 2022. PMID: 36356384 Review.
Cited by
-
Label-free morpho-molecular phenotyping of living cancer cells by combined Raman spectroscopy and phase tomography.Commun Biol. 2024 Jun 29;7(1):785. doi: 10.1038/s42003-024-06496-9. Commun Biol. 2024. PMID: 38951178 Free PMC article.
-
In-silico and in-vitro morphometric analysis of intestinal organoids.PLoS Comput Biol. 2023 Aug 14;19(8):e1011386. doi: 10.1371/journal.pcbi.1011386. eCollection 2023 Aug. PLoS Comput Biol. 2023. PMID: 37578984 Free PMC article.
-
GEMA-An Automatic Segmentation Method for Real-Time Analysis of Mammalian Cell Growth in Microfluidic Devices.J Imaging. 2022 Oct 14;8(10):281. doi: 10.3390/jimaging8100281. J Imaging. 2022. PMID: 36286375 Free PMC article.
-
Virtual labeling of mitochondria in living cells using correlative imaging and physics-guided deep learning.Biomed Opt Express. 2022 Sep 28;13(10):5495-5516. doi: 10.1364/BOE.464177. eCollection 2022 Oct 1. Biomed Opt Express. 2022. PMID: 36425635 Free PMC article.
-
Quantitative phase velocimetry measures bulk intracellular transport of cell mass during the cell cycle.Sci Rep. 2022 Apr 12;12(1):6074. doi: 10.1038/s41598-022-10000-w. Sci Rep. 2022. PMID: 35414087 Free PMC article.
References
-
- Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng. 2010;57(4):841–52. - PubMed
-
- Dimopoulos S, Mayer CE, Rudolf F, Stelling J. Accurate cell segmentation in microscopy images using membrane patterns, Bioinformatics (Oxford, England) 2014;30(18):2644–51. - PubMed
-
- Hilsenbeck O, Schwarzfischer M, Loeffler D, Dimopoulos S, Hastreiter S, Marr C, Theis FJ, Schroeder T. Faster: a user-friendly tool for ultrafast and robust cell segmentation in large-scale microscopy. Bioinformatics. 2017;33(13):2020–28. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

