Decoding the proteomic changes involved in the biofilm formation of Enterococcus faecalis SK460 to elucidate potential biofilm determinants
- PMID: 31253082
- PMCID: PMC6599329
- DOI: 10.1186/s12866-019-1527-2
Decoding the proteomic changes involved in the biofilm formation of Enterococcus faecalis SK460 to elucidate potential biofilm determinants
Abstract
Background: Enterococcus faecalis is a major clinically relevant nosocomial bacterial pathogen frequently isolated from polymicrobial infections. The biofilm forming ability of E. faecalis attributes a key role in its virulence and drug resistance. Biofilm cells are phenotypically and metabolically different from their planktonic counterparts and many aspects involved in E. faecalis biofilm formation are yet to be elucidated. The strain E. faecalis SK460 used in the present study is esp (Enterococcal surface protein) and fsr (two-component signal transduction system) negative non-gelatinase producing strong biofilm former isolated from a chronic diabetic foot ulcer patient. We executed a label-free quantitative proteomic approach to elucidate the differential protein expression pattern at planktonic and biofilm stages of SK460 to come up with potential determinants associated with Enterococcal biofilm formation.
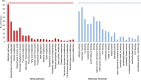
Results: The Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of proteomic data revealed that biofilm cells expressed higher levels of proteins which are associated with glycolysis, amino acid biosynthesis, biosynthesis of secondary metabolites, microbial metabolism in diverse environments and stress response factors. Besides these basic survival pathways, LuxS-mediated quorum sensing, arginine metabolism, rhamnose biosynthesis, pheromone and adhesion associated proteins were found to be upregulated during the biofilm transit from planktonic stages. The selected subsets were validated by quantitative real-time PCR. In silico functional interaction analysis revealed that the genes involved in upregulated pathways pose a close molecular interaction thereby coordinating the regulatory network to thrive as a biofilm community.
Conclusions: The present study describes the first report of the quantitative proteome analysis of an esp and fsr negative non gelatinase producing E. faecalis. Proteome analysis evidenced enhanced expression of glycolytic pathways, stress response factors, LuxS quorum signaling system, rhamnopolysaccharide synthesis and pheromone associated proteins in biofilm phenotype. We also pointed out the relevance of LuxS quorum sensing and pheromone associated proteins in the biofilm development of E. faecalis which lacks the Fsr quorum signaling system. These validated biofilm determinants can act as potential inhibiting targets in Enterococcal infections.
Keywords: Biofilm determinants; Enterococcus faecalis; Metabolic pathways; Quantitative proteomics; Stress response; luxS.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
LuxS-dependent AI-2 regulates versatile functions in Enterococcus faecalis V583.J Proteome Res. 2012 Sep 7;11(9):4465-75. doi: 10.1021/pr3002244. Epub 2012 Aug 15. J Proteome Res. 2012. PMID: 22856334
-
Molecular Mechanism of Quorum-Sensing in Enterococcus faecalis: Its Role in Virulence and Therapeutic Approaches.Int J Mol Sci. 2017 May 3;18(5):960. doi: 10.3390/ijms18050960. Int J Mol Sci. 2017. PMID: 28467378 Free PMC article. Review.
-
Siamycin attenuates fsr quorum sensing mediated by a gelatinase biosynthesis-activating pheromone in Enterococcus faecalis.J Bacteriol. 2007 Feb;189(4):1358-65. doi: 10.1128/JB.00969-06. Epub 2006 Oct 27. J Bacteriol. 2007. PMID: 17071762 Free PMC article.
-
Polidocanol inhibits Enterococcus faecalis virulence factors by targeting fsr quorum sensing system.BMC Microbiol. 2024 Oct 16;24(1):411. doi: 10.1186/s12866-024-03548-2. BMC Microbiol. 2024. PMID: 39415105 Free PMC article.
-
An Overview of the Factors Involved in Biofilm Production by the Enterococcus Genus.Int J Mol Sci. 2023 Jul 18;24(14):11577. doi: 10.3390/ijms241411577. Int J Mol Sci. 2023. PMID: 37511337 Free PMC article. Review.
Cited by
-
How to study biofilms: technological advancements in clinical biofilm research.Front Cell Infect Microbiol. 2023 Dec 13;13:1335389. doi: 10.3389/fcimb.2023.1335389. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38156318 Free PMC article. Review.
-
Triple threat: how diabetes results in worsened bacterial infections.Infect Immun. 2024 Sep 10;92(9):e0050923. doi: 10.1128/iai.00509-23. Epub 2024 Mar 25. Infect Immun. 2024. PMID: 38526063 Free PMC article. Review.
-
An Overview of Biological and Computational Methods for Designing Mechanism-Informed Anti-biofilm Agents.Front Microbiol. 2021 Apr 13;12:640787. doi: 10.3389/fmicb.2021.640787. eCollection 2021. Front Microbiol. 2021. PMID: 33927701 Free PMC article. Review.
-
The de novo Purine Biosynthesis Pathway Is the Only Commonly Regulated Cellular Pathway during Biofilm Formation in TSB-Based Medium in Staphylococcus aureus and Enterococcus faecalis.Microbiol Spectr. 2021 Dec 22;9(3):e0080421. doi: 10.1128/Spectrum.00804-21. Epub 2021 Dec 22. Microbiol Spectr. 2021. PMID: 34935415 Free PMC article.
-
Genome-resolved metagenomics provides insights into the ecological roles of the keystone taxa in heavy-metal-contaminated soils.Front Microbiol. 2023 Jul 21;14:1203164. doi: 10.3389/fmicb.2023.1203164. eCollection 2023. Front Microbiol. 2023. PMID: 37547692 Free PMC article.
References
-
- Bourgogne A, Singh KV, Fox KA, Pflughoeft KJ, Murray BE, Garsin DA. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J Bacteriol. 2007;189(17):6490–6493. doi: 10.1128/JB.00594-07. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

