Construction and analysis of degradome-dependent microRNA regulatory networks in soybean
- PMID: 31253085
- PMCID: PMC6599275
- DOI: 10.1186/s12864-019-5879-7
Construction and analysis of degradome-dependent microRNA regulatory networks in soybean
Abstract
Background: Usually the microRNA (miRNA)-mediated gene regulatory network (GRN) is constructed from the investigation of miRNA expression profiling and target predictions. However, the higher/lower expression level of miRNAs does not always indicate the higher/lower level of cleavages and such analysis, thus, sometimes ignores the crucial cleavage events. In the present work, the degradome sequencing data were employed to construct the complete miRNA-mediated gene regulatory network in soybean, unlike the traditional approach starting with small RNA sequencing data.
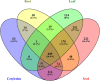
Results: We constructed the root-, cotyledon-, leaf- and seed-specific miRNA regulatory networks with the degradome sequencing data and the forthcoming verification of miRNA profiling analysis. As a result, we identified 205 conserved miRNA-target interactions (MTIs) involved with 6 conserved gma-miRNA families and 365 tissue-specific MTIs containing 24 root-specific, 45 leaf-specific, 63 cotyledon-specific and 225 seed-specific MTIs. We found a total of 156 miRNAs in tissue-specific MTIs including 18 tissue-specific miRNAs, however, only 3 miRNAs have consistent tissue-specific expression. Our study showed the degradome-dependent miRNA regulatory networks (DDNs) in four soybean tissues and explored their conservations and specificities.
Conclusions: The construction of DDNs may provide the complete miRNA-Target interactions in certain plant tissues, leading to the identification of the conserved and tissue-specific MTIs and sub-networks. Our work provides a basis for further investigation of the roles and mechanisms of miRNA-mediated regulation of tissue-specific growth and development in soybean.
Keywords: DDN; Degradome; Regulatory network; Soybean; microRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing.BMC Genomics. 2012 Jul 16;13:310. doi: 10.1186/1471-2164-13-310. BMC Genomics. 2012. PMID: 22799740 Free PMC article.
-
Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing.BMC Plant Biol. 2011 Jan 10;11:5. doi: 10.1186/1471-2229-11-5. BMC Plant Biol. 2011. PMID: 21219599 Free PMC article.
-
Analyses of a Glycine max degradome library identify microRNA targets and microRNAs that trigger secondary siRNA biogenesis.J Integr Plant Biol. 2013 Feb;55(2):160-76. doi: 10.1111/jipb.12002. J Integr Plant Biol. 2013. PMID: 23131131
-
The use of high-throughput sequencing methods for plant microRNA research.RNA Biol. 2015;12(7):709-19. doi: 10.1080/15476286.2015.1053686. RNA Biol. 2015. PMID: 26016494 Free PMC article. Review.
-
miRNA degradation in the mammalian brain.Am J Physiol Cell Physiol. 2020 Oct 1;319(4):C624-C629. doi: 10.1152/ajpcell.00303.2020. Epub 2020 Aug 12. Am J Physiol Cell Physiol. 2020. PMID: 32783657 Free PMC article. Review.
Cited by
-
Origin and Evolutionary Dynamics of the miR2119 and ADH1 Regulatory Module in Legumes.Genome Biol Evol. 2020 Dec 6;12(12):2355-2369. doi: 10.1093/gbe/evaa205. Genome Biol Evol. 2020. PMID: 33045056 Free PMC article.
-
A Tissue-Specific Landscape of Alternative Polyadenylation, lncRNAs, TFs, and Gene Co-expression Networks in Liriodendron chinense.Front Plant Sci. 2021 Jul 23;12:705321. doi: 10.3389/fpls.2021.705321. eCollection 2021. Front Plant Sci. 2021. PMID: 34367224 Free PMC article.
-
A Multi-Level Iterative Bi-Clustering Method for Discovering miRNA Co-regulation Network of Abiotic Stress Tolerance in Soybeans.Front Plant Sci. 2022 Apr 7;13:860791. doi: 10.3389/fpls.2022.860791. eCollection 2022. Front Plant Sci. 2022. PMID: 35463453 Free PMC article.
-
Integrative expression network analysis of microRNA and gene isoforms in sacred lotus.BMC Genomics. 2020 Jun 25;21(1):429. doi: 10.1186/s12864-020-06853-y. BMC Genomics. 2020. PMID: 32586276 Free PMC article.
-
Non-Coding RNAs in Legumes: Their Emerging Roles in Regulating Biotic/Abiotic Stress Responses and Plant Growth and Development.Cells. 2021 Jul 2;10(7):1674. doi: 10.3390/cells10071674. Cells. 2021. PMID: 34359842 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

