Odyssey: a semi-automated pipeline for phasing, imputation, and analysis of genome-wide genetic data
- PMID: 31253090
- PMCID: PMC6599316
- DOI: 10.1186/s12859-019-2964-5
Odyssey: a semi-automated pipeline for phasing, imputation, and analysis of genome-wide genetic data
Abstract
Background: Genome imputation, admixture resolution and genome-wide association analyses are timely and computationally intensive processes with many composite and requisite steps. Analysis time increases further when building and installing the run programs required for these analyses. For scientists that may not be as versed in programing language, but want to perform these operations hands on, there is a lengthy learning curve to utilize the vast number of programs available for these analyses.
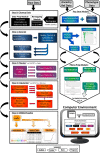
Results: In an effort to streamline the entire process with easy-to-use steps for scientists working with big data, the Odyssey pipeline was developed. Odyssey is a simplified, efficient, semi-automated genome-wide imputation and analysis pipeline, which prepares raw genetic data, performs pre-imputation quality control, phasing, imputation, post-imputation quality control, population stratification analysis, and genome-wide association with statistical data analysis, including result visualization. Odyssey is a pipeline that integrates programs such as PLINK, SHAPEIT, Eagle, IMPUTE, Minimac, and several R packages, to create a seamless, easy-to-use, and modular workflow controlled via a single user-friendly configuration file. Odyssey was built with compatibility in mind, and thus utilizes the Singularity container solution, which can be run on Linux, MacOS, and Windows platforms. It is also easily scalable from a simple desktop to a High-Performance System (HPS).
Conclusion: Odyssey facilitates efficient and fast genome-wide association analysis automation and can go from raw genetic data to genome: phenome association visualization and analyses results in 3-8 h on average, depending on the input data, choice of programs within the pipeline and available computer resources. Odyssey was built to be flexible, portable, compatible, scalable, and easy to setup. Biologists less familiar with programing can now work hands on with their own big data using this easy-to-use pipeline.
Keywords: Admixture; Genome-wide-association study; Imputation; Odyssey; Phasing; Pipeline.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
ILIAD: a suite of automated Snakemake workflows for processing genomic data for downstream applications.BMC Bioinformatics. 2023 Nov 8;24(1):424. doi: 10.1186/s12859-023-05548-x. BMC Bioinformatics. 2023. PMID: 37940870 Free PMC article.
-
Molgenis-impute: imputation pipeline in a box.BMC Res Notes. 2015 Aug 19;8:359. doi: 10.1186/s13104-015-1309-3. BMC Res Notes. 2015. PMID: 26286716 Free PMC article.
-
Gimpute: an efficient genetic data imputation pipeline.Bioinformatics. 2019 Apr 15;35(8):1433-1435. doi: 10.1093/bioinformatics/bty814. Bioinformatics. 2019. PMID: 30239591
-
Genotype imputation in genome-wide association studies.Curr Protoc Hum Genet. 2013 Jul;Chapter 1:Unit 1.25. doi: 10.1002/0471142905.hg0125s78. Curr Protoc Hum Genet. 2013. PMID: 23853078 Review.
-
A Pipeline for Phasing and Genotype Imputation on Mixed Human Data (Parents-Offspring Trios and Unrelated Subjects) by Reviewing Current Methods and Software.Life (Basel). 2022 Dec 5;12(12):2030. doi: 10.3390/life12122030. Life (Basel). 2022. PMID: 36556394 Free PMC article. Review.
Cited by
-
ILIAD: a suite of automated Snakemake workflows for processing genomic data for downstream applications.BMC Bioinformatics. 2023 Nov 8;24(1):424. doi: 10.1186/s12859-023-05548-x. BMC Bioinformatics. 2023. PMID: 37940870 Free PMC article.
-
Shared heritability of human face and brain shape.Nat Genet. 2021 Jun;53(6):830-839. doi: 10.1038/s41588-021-00827-w. Epub 2021 Apr 5. Nat Genet. 2021. PMID: 33821002 Free PMC article.
-
Canary: an automated tool for the conversion of MaCH imputed dosage files to PLINK files.BMC Bioinformatics. 2022 Jul 27;23(1):304. doi: 10.1186/s12859-022-04822-8. BMC Bioinformatics. 2022. PMID: 35896971 Free PMC article.
-
BIGwas: Single-command quality control and association testing for multi-cohort and biobank-scale GWAS/PheWAS data.Gigascience. 2021 Jun 29;10(6):giab047. doi: 10.1093/gigascience/giab047. Gigascience. 2021. PMID: 34184051 Free PMC article.
-
nf-gwas-pipeline: A Nextflow Genome-Wide Association Study Pipeline.J Open Source Softw. 2021;6(59):2957. doi: 10.21105/joss.02957. Epub 2021 Mar 2. J Open Source Softw. 2021. PMID: 35647481 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

