Genome Editing of Expanded CTG Repeats within the Human DMPK Gene Reduces Nuclear RNA Foci in the Muscle of DM1 Mice
- PMID: 31253581
- PMCID: PMC6697452
- DOI: 10.1016/j.ymthe.2019.05.021
Genome Editing of Expanded CTG Repeats within the Human DMPK Gene Reduces Nuclear RNA Foci in the Muscle of DM1 Mice
Abstract
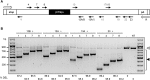
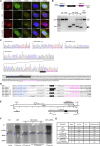
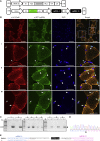
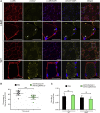
Myotonic dystrophy type 1 (DM1) is caused by a CTG repeat expansion located in the 3' UTR of the DMPK gene. Expanded DMPK transcripts aggregate into nuclear foci and alter the function of RNA-binding proteins, leading to defects in the alternative splicing of numerous pre-mRNAs. To date, there is no curative treatment for DM1. Here we investigated a gene-editing strategy using the CRISPR-Cas9 system from Staphylococcus aureus (Sa) to delete the CTG repeats in the human DMPK locus. Co-expression of SaCas9 and selected pairs of single-guide RNAs (sgRNAs) in cultured DM1 patient-derived muscle line cells carrying 2,600 CTG repeats resulted in targeted DNA deletion, ribonucleoprotein foci disappearance, and correction of splicing abnormalities in various transcripts. Furthermore, a single intramuscular injection of recombinant AAV vectors expressing CRISPR-SaCas9 components in the tibialis anterior muscle of DMSXL (myotonic dystrophy mouse line carrying the human DMPK gene with >1,000 CTG repeats) mice decreased the number of pathological RNA foci in myonuclei. These results establish the proof of concept that genome editing of a large trinucleotide expansion is feasible in muscle and may represent a useful strategy to be further developed for the treatment of myotonic dystrophy.
Keywords: CRISPR-Cas9; DMPK; gene therapy; myotonic dystrophy; nucleotide repeat disorders.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Udd B., Krahe R. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012;11:891–905. - PubMed
-
- Buxton J., Shelbourne P., Davies J., Jones C., Van Tongeren T., Aslanidis C., de Jong P., Jansen G., Anvret M., Riley B. Detection of an unstable fragment of DNA specific to individuals with myotonic dystrophy. Nature. 1992;355:547–548. - PubMed
-
- Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. - PubMed
-
- Harley H.G., Brook J.D., Rundle S.A., Crow S., Reardon W., Buckler A.J., Harper P.S., Housman D.E., Shaw D.J. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature. 1992;355:545–546. - PubMed
-
- Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O’Hoy K. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

