A Global Survey of Mycobacterial Diversity in Soil
- PMID: 31253672
- PMCID: PMC6696970
- DOI: 10.1128/AEM.01180-19
A Global Survey of Mycobacterial Diversity in Soil
Abstract
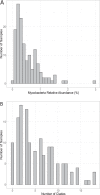
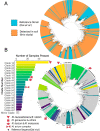
Mycobacteria are a diverse bacterial group ubiquitous in many soil and aquatic environments. Members of this group have been associated with human and other animal diseases, including the nontuberculous mycobacteria (NTM), which are of growing relevance to public health worldwide. Although soils are often considered an important source of environmentally acquired NTM infections, the biodiversity and ecological preferences of soil mycobacteria remain largely unexplored across contrasting climates and ecosystem types. Using a culture-independent approach by combining 16S rRNA marker gene sequencing with mycobacterium-specific hsp65 gene sequencing, we analyzed the diversity, distributions, and environmental preferences of soil-dwelling mycobacteria in 143 soil samples collected from a broad range of ecosystem types. The surveyed soils harbored highly diverse mycobacterial communities that span the full extent of the known mycobacterial phylogeny, with most soil mycobacteria (97% of mycobacterial clades) belonging to previously undescribed lineages. While mycobacteria tended to have higher relative abundances in cool, wet, and acidic soil environments, several individual mycobacterial clades had contrasting environmental preferences. We identified the environmental preferences of many mycobacterial clades, including the clinically relevant Mycobacterium avium complex that was more commonly detected in wet and acidic soils. However, most of the soil mycobacteria detected were not closely related to known pathogens, calling into question previous assumptions about the general importance of soil as a source of NTM infections. Together, this work provides novel insights into the diversity, distributions, and ecological preferences of soil mycobacteria and lays the foundation for future efforts to link mycobacterial phenotypes to their distributions.IMPORTANCE Mycobacteria are common inhabitants of soil, and while most members of this bacterial group are innocuous, some mycobacteria can cause environmentally acquired infections of humans and other animals. Human infections from nontuberculous mycobacteria (NTM) are increasingly prevalent worldwide, and some areas appear to be "hotspots" for NTM disease. While exposure to soil is frequently implicated as an important mode of NTM transmission, the diversity, distributions, and ecological preferences of soil mycobacteria remain poorly understood. We analyzed 143 soils from a range of ecosystems and found that mycobacteria and lineages within the group often exhibited predictable preferences for specific environmental conditions. Soils harbor large amounts of previously undescribed mycobacterial diversity, and lineages that include known pathogens were rarely detected in soil. Together, these findings suggest that soil is an unlikely source of many mycobacterial infections. The biogeographical patterns we documented lend insight into the ecology of this important group of soil-dwelling bacteria.
Keywords: Mycobacterium; mycobacteria; soil microbiology.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Hruska K, Kaevska M. 2013. Mycobacteria in water, soil, plants and air: a review. Veterinarni Medicina 57:623–679. doi:10.17221/6558-VETMED. - DOI
-
- Tortoli E. 2019. The taxonomy of the genus Mycobacterium, p 1–10. In Velayati AA, Farnia P (ed), Nontuberculous mycobacteria (NTM). Academic Press, Cambridge, MA. doi:10.1016/B978-0-12-814692-7.00001-2. - DOI
-
- Parte AC. 2018. LPSN—List of Prokaryotic Names with Standing in Nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol 68:1825–1829. doi:10.1099/ijsem.0.002786. - DOI - PubMed
-
- Hennessee CT, Seo J-S, Alvarez AM, Li QX. 2009. Polycyclic aromatic hydrocarbon-degrading species isolated from Hawaiian soils: Mycobacterium crocinum sp. nov., Mycobacterium pallens sp. nov., Mycobacterium rutilum sp. nov., Mycobacterium rufum sp. nov. and Mycobacterium aromaticivorans sp. nov. Int J Syst Evol Microbiol 59:378–387. doi:10.1099/ijs.0.65827-0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

