Unexpected host dependency of Antarctic Nanohaloarchaeota
- PMID: 31253704
- PMCID: PMC6642349
- DOI: 10.1073/pnas.1905179116
Unexpected host dependency of Antarctic Nanohaloarchaeota
Abstract
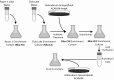
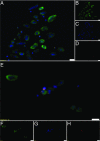
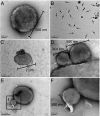
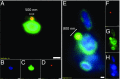
In hypersaline environments, Nanohaloarchaeota (Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, Nanohaloarchaeota [DPANN] superphylum) are thought to be free-living microorganisms. We report cultivation of 2 strains of Antarctic Nanohaloarchaeota and show that they require the haloarchaeon Halorubrum lacusprofundi for growth. By performing growth using enrichments and fluorescence-activated cell sorting, we demonstrated successful cultivation of Candidatus Nanohaloarchaeum antarcticus, purification of Ca. Nha. antarcticus away from other species, and growth and verification of Ca. Nha. antarcticus with Hrr. lacusprofundi; these findings are analogous to those required for fulfilling Koch's postulates. We use fluorescent in situ hybridization and transmission electron microscopy to assess cell structures and interactions; metagenomics to characterize enrichment taxa, generate metagenome assembled genomes, and interrogate Antarctic communities; and proteomics to assess metabolic pathways and speculate about the roles of certain proteins. Metagenome analysis indicates the presence of a single species, which is endemic to Antarctic hypersaline systems that support the growth of haloarchaea. The presence of unusually large proteins predicted to function in attachment and invasion of hosts plus the absence of key biosynthetic pathways (e.g., lipids) in metagenome assembled genomes of globally distributed Nanohaloarchaeota indicate that all members of the lineage have evolved as symbionts. Our work expands the range of archaeal symbiotic lifestyles and provides a genetically tractable model system for advancing understanding of the factors controlling microbial symbiotic relationships.
Keywords: DPANN; archaea; symbiont.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Spang A., Caceres E. F., Ettema T. J. G., Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science 357, eaaf3883 (2017). - PubMed
-
- Rinke C., et al. , Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013). - PubMed

