Structural basis of TFIIH activation for nucleotide excision repair
- PMID: 31253769
- PMCID: PMC6599211
- DOI: 10.1038/s41467-019-10745-5
Structural basis of TFIIH activation for nucleotide excision repair
Abstract
Nucleotide excision repair (NER) is the major DNA repair pathway that removes UV-induced and bulky DNA lesions. There is currently no structure of NER intermediates, which form around the large multisubunit transcription factor IIH (TFIIH). Here we report the cryo-EM structure of an NER intermediate containing TFIIH and the NER factor XPA. Compared to its transcription conformation, the TFIIH structure is rearranged such that its ATPase subunits XPB and XPD bind double- and single-stranded DNA, consistent with their translocase and helicase activities, respectively. XPA releases the inhibitory kinase module of TFIIH, displaces a 'plug' element from the DNA-binding pore in XPD, and together with the NER factor XPG stimulates XPD activity. Our results explain how TFIIH is switched from a transcription to a repair factor, and provide the basis for a mechanistic analysis of the NER pathway.
Conflict of interest statement
The authors declare no competing interests.
Figures

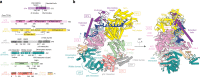


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

