Mutant H3 histones drive human pre-leukemic hematopoietic stem cell expansion and promote leukemic aggressiveness
- PMID: 31253791
- PMCID: PMC6599207
- DOI: 10.1038/s41467-019-10705-z
Mutant H3 histones drive human pre-leukemic hematopoietic stem cell expansion and promote leukemic aggressiveness
Abstract
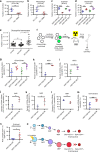
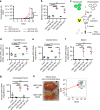
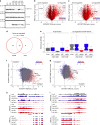
Our ability to manage acute myeloid leukemia (AML) is limited by our incomplete understanding of the epigenetic disruption central to leukemogenesis, including improper histone methylation. Here we examine 16 histone H3 genes in 434 primary AML samples and identify Q69H, A26P, R2Q, R8H and K27M/I mutations (1.6%), with higher incidence in secondary AML (9%). These mutations occur in pre-leukemic hematopoietic stem cells (HSCs) and exist in the major leukemic clones in patients. They increase the frequency of functional HSCs, alter differentiation, and amplify leukemic aggressiveness. These effects are dependent on the specific mutation. H3K27 mutation increases the expression of genes involved in erythrocyte and myeloid differentiation with altered H3K27 tri-methylation and K27 acetylation. The functional impact of histone mutations is independent of RUNX1 mutation, although they at times co-occur. This study establishes that H3 mutations are drivers of human pre-cancerous stem cell expansion and important early events in leukemogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

