Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes
- PMID: 31253982
- PMCID: PMC6640048
- DOI: 10.1038/s41588-019-0457-0
Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes
Abstract
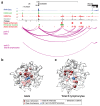
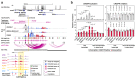
Genetic studies promise to provide insight into the molecular mechanisms underlying type 2 diabetes (T2D). Variants associated with T2D are often located in tissue-specific enhancer clusters or super-enhancers. So far, such domains have been defined through clustering of enhancers in linear genome maps rather than in three-dimensional (3D) space. Furthermore, their target genes are often unknown. We have created promoter capture Hi-C maps in human pancreatic islets. This linked diabetes-associated enhancers to their target genes, often located hundreds of kilobases away. It also revealed >1,300 groups of islet enhancers, super-enhancers and active promoters that form 3D hubs, some of which show coordinated glucose-dependent activity. We demonstrate that genetic variation in hubs impacts insulin secretion heritability, and show that hub annotations can be used for polygenic scores that predict T2D risk driven by islet regulatory variants. Human islet 3D chromatin architecture, therefore, provides a framework for interpretation of T2D genome-wide association study (GWAS) signals.
Conflict of interest statement
P. R. is a shareholder and consultant for Endocells/Unicercell Biosolutions.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

