A recommended and verified procedure for in situ tryptic digestion of formalin-fixed paraffin-embedded tissues for analysis by matrix-assisted laser desorption/ionization imaging mass spectrometry
- PMID: 31254303
- PMCID: PMC6711785
- DOI: 10.1002/jms.4384
A recommended and verified procedure for in situ tryptic digestion of formalin-fixed paraffin-embedded tissues for analysis by matrix-assisted laser desorption/ionization imaging mass spectrometry
Abstract
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) is a molecular imaging technology uniquely capable of untargeted measurement of proteins, lipids, and metabolites while retaining spatial information about their location in situ. This powerful combination of capabilities has the potential to bring a wealth of knowledge to the field of molecular histology. Translation of this innovative research tool into clinical laboratories requires the development of reliable sample preparation protocols for the analysis of proteins from formalin-fixed paraffin-embedded (FFPE) tissues, the standard preservation process in clinical pathology. Although ideal for stained tissue analysis by microscopy, the FFPE process cross-links, disrupts, or can remove proteins from the tissue, making analysis of the protein content challenging. To date, reported approaches differ widely in process and efficacy. This tutorial presents a strategy derived from systematic testing and optimization of key parameters, for reproducible in situ tryptic digestion of proteins in FFPE tissue and subsequent MALDI IMS analysis. The approach describes a generalized method for FFPE tissues originating from virtually any source.
Keywords: MALDI imaging mass spectrometry; colon; formalin-fixed paraffin-embedded tissue; in situ tryptic digestion; molecular histology.
© 2019 John Wiley & Sons, Ltd.
Figures

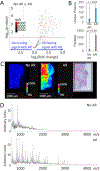

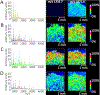




Similar articles
-
On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry.J Am Soc Mass Spectrom. 2010 Mar;21(3):338-47. doi: 10.1016/j.jasms.2009.09.016. Epub 2009 Sep 29. J Am Soc Mass Spectrom. 2010. PMID: 19926301
-
Tissue fixed with formalin and processed without paraffin embedding is suitable for imaging of both peptides and lipids by MALDI-IMS.Proteomics. 2016 Jun;16(11-12):1670-7. doi: 10.1002/pmic.201500424. Epub 2016 May 25. Proteomics. 2016. PMID: 27001204
-
MALDI Imaging Mass Spectrometry of N-glycans and Tryptic Peptides from the Same Formalin-Fixed, Paraffin-Embedded Tissue Section.Methods Mol Biol. 2018;1788:225-241. doi: 10.1007/7651_2017_81. Methods Mol Biol. 2018. PMID: 29058228 Free PMC article.
-
MALDI mass spectrometry imaging of formalin-fixed paraffin-embedded tissues in clinical research.Histol Histopathol. 2014 Nov;29(11):1365-76. doi: 10.14670/HH-29.1365. Epub 2014 May 19. Histol Histopathol. 2014. PMID: 24838644 Review.
-
MALDI IMS and Cancer Tissue Microarrays.Adv Cancer Res. 2017;134:173-200. doi: 10.1016/bs.acr.2016.11.007. Epub 2017 Jan 12. Adv Cancer Res. 2017. PMID: 28110650 Review.
Cited by
-
Helicobacter pylori CagA and Cag type IV secretion system activity have key roles in triggering gastric transcriptional and proteomic alterations.Infect Immun. 2025 Apr 8;93(4):e0059524. doi: 10.1128/iai.00595-24. Epub 2025 Mar 6. Infect Immun. 2025. PMID: 40047510 Free PMC article.
-
MALDI-MSI as a Complementary Diagnostic Tool in Cytopathology: A Pilot Study for the Characterization of Thyroid Nodules.Cancers (Basel). 2019 Sep 16;11(9):1377. doi: 10.3390/cancers11091377. Cancers (Basel). 2019. PMID: 31527543 Free PMC article.
-
Bottom-Up Proteomics: Advancements in Sample Preparation.Int J Mol Sci. 2023 Mar 10;24(6):5350. doi: 10.3390/ijms24065350. Int J Mol Sci. 2023. PMID: 36982423 Free PMC article. Review.
-
Remodeling of the gastric environment in Helicobacter pylori-induced atrophic gastritis.mSystems. 2024 Jan 23;9(1):e0109823. doi: 10.1128/msystems.01098-23. Epub 2023 Dec 7. mSystems. 2024. PMID: 38059647 Free PMC article.
-
A simple preparation protocol for shipping and storage of tissue sections for laser ablation-inductively coupled plasma-mass spectrometry imaging.Metallomics. 2022 Mar 28;14(3):mfac013. doi: 10.1093/mtomcs/mfac013. Metallomics. 2022. PMID: 35294013 Free PMC article.
References
-
- Caprioli RM, Farmer TB, and Gile J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem 1997, 69, 4751. - PubMed
-
- Ly A, Buck A, Balluff B, Sun N, Gorzolka K, Feuchtinger A, Janssen K-P, Kuppen PJK, van de Velde CJH, Weirich G, Erlmeier F, Langer R, Aubele M, Zitzelsberger H, McDonnell L, Aichler M, and Walch A. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nature Protocols 2016, 11, 1428. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

