Universality and Structural Implications of the Boson Peak in Proteins
- PMID: 31255295
- PMCID: PMC6700671
- DOI: 10.1016/j.bpj.2019.06.007
Universality and Structural Implications of the Boson Peak in Proteins
Abstract
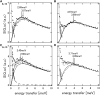
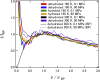
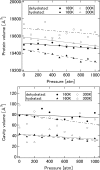
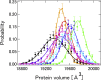
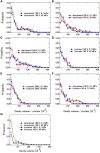
The softness and rigidity of proteins are reflected in the structural dynamics, which are in turn affected by the environment. The characteristic low-frequency vibrational spectrum of a protein, known as boson peak, is an indication of the structural rigidity of the protein at a cryogenic temperature or dehydrated conditions. In this article, the effect of hydration, temperature, and pressure on the boson peak and volumetric properties of a globular protein are evaluated by using inelastic neutron scattering and molecular dynamics simulation. Hydration, pressurization, and cooling shift the boson peak position to higher energy and depress the peak intensity and decreases the protein and cavity volumes. We found the correlation between the boson peak and cavity volume in a protein. A decrease of cavity volume means the increase of rigidity, which is the origin of the boson peak shift. Boson peak is the universal property of a protein, which is rationalized by the correlation.
Copyright © 2019 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Doster W., Cusack S., Petry W. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature. 1989;337:754–756. - PubMed
-
- Zaccai G. How soft is a protein? A protein dynamics force constant measured by neutron scattering. Science. 2000;288:1604–1607. - PubMed
-
- Rasmussen B.F., Stock A.M., Petsko G.A. Crystalline ribonuclease A loses function below the dynamical transition at 220 K. Nature. 1992;357:423–424. - PubMed
-
- Doster W., Settles M. Protein-water displacement distributions. Biochim. Biophys. Acta. 2005;1749:173–186. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

