Development and Clinical Validation of a Large Fusion Gene Panel for Pediatric Cancers
- PMID: 31255796
- PMCID: PMC6734859
- DOI: 10.1016/j.jmoldx.2019.05.006
Development and Clinical Validation of a Large Fusion Gene Panel for Pediatric Cancers
Abstract
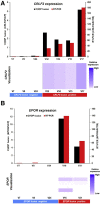
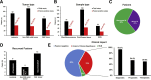
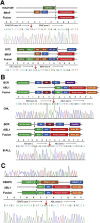
Gene fusions are one of the most common genomic alterations in pediatric cancer. Many fusions encode oncogenic drivers and play important roles in cancer diagnosis, risk stratification, and treatment selection. We report the development and clinical validation of a large custom-designed RNA sequencing panel, CHOP Fusion panel, using anchored multiplex PCR technology. The panel interrogates 106 cancer genes known to be involved in nearly 600 different fusions reported in hematological malignancies and solid tumors. The panel works well with different types of samples, including formalin-fixed, paraffin-embedded samples. The panel demonstrated excellent analytic accuracy, with 100% sensitivity and specificity on 60 pediatric tumor validation samples. In addition to identifying all known fusions in the validation samples, three unrecognized, yet clinically significant, fusions were also detected. A total of 276 clinical cases were analyzed after the validation, and 51 different fusions were identified in 104 cases. Of these fusions, 16 were not previously reported at the time of discovery. These fusions provided genomic information useful for clinical management. Our experience demonstrates that CHOP Fusion panel can detect the vast majority of known and certain novel clinically relevant fusion genes in pediatric cancers accurately, efficiently, and cost-effectively; and the panel provides an excellent tool for new fusion gene discovery.
Copyright © 2019 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. - PubMed
-
- Mertens F., Johansson B., Fioretos T., Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. - PubMed
-
- Teixeira M.R. Recurrent fusion oncogenes in carcinomas. Crit Rev Oncog. 2006;12:257–271. - PubMed
-
- Li M.M., Ewton A.A., Smith J.L. Using cytogenetic rearrangements for cancer prognosis and treatment (pharmacogenetics) Curr Genet Med Rep. 2013;1:99–112.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

