PPR-SMR1 is required for the splicing of multiple mitochondrial introns, interacts with Zm-mCSF1, and is essential for seed development in maize
- PMID: 31257441
- PMCID: PMC6793435
- DOI: 10.1093/jxb/erz305
PPR-SMR1 is required for the splicing of multiple mitochondrial introns, interacts with Zm-mCSF1, and is essential for seed development in maize
Abstract
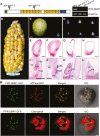
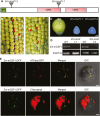
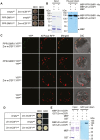
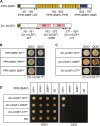
Group II introns are ribozymes that can excise themselves from precursor-RNA transcripts, but plant organellar group II introns have structural deviations that inhibit ribozyme activity. Therefore, splicing of these introns requires the assistance of nuclear- and/or organellar-encoded splicing factors; however, how these splicing factors function remains unclear. In this study, we report the functions and interactions of two splicing factors, PPR-SMR1 and Zm-mCSF1, in intron splicing in maize mitochondria. PPR-SMR1 is a SMR domain-containing pentatricopeptide repeat (PPR) protein and Zm-mCSF1 is a CRM domain-containing protein, and both are targeted to mitochondria. Loss-of-function mutations in each of them severely arrests embryogenesis and endosperm development in maize. Functional analyses indicate that PPR-SMR1 and Zm-mCSF1 are required for the splicing of most mitochondrial group II introns. Among them, nad2-intron 2 and 3, and nad5-intron 1 are PPR-SMR1/Zm-mCSF1-dependent introns. Protein interaction assays suggest that PPR-SMR1 can interact with Zm-mCSF1 through its N-terminus, and that Zm-mCSF1 is self-interacting. Our findings suggest that PPR-SMR1, a novel splicing factor, acts in the splicing of multiple group II introns in maize mitochondria, and the protein-protein interaction between it and Zm-mCSF1 might allow the formation of large macromolecular splicing complexes.
Keywords: Group II introns; maize; mitochondria; organelle biogenesis; pentatricopeptide repeat (PPR) proteins; seed development.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

