Splicing dysregulation contributes to the pathogenicity of several F9 exonic point variants
- PMID: 31257730
- PMCID: PMC6687662
- DOI: 10.1002/mgg3.840
Splicing dysregulation contributes to the pathogenicity of several F9 exonic point variants
Abstract
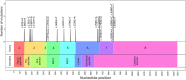
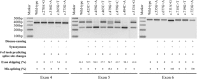
Background: Pre-mRNA splicing is a complex process requiring the identification of donor site, acceptor site, and branch point site with an adjacent polypyrimidine tract sequence. Splicing is regulated by splicing regulatory elements (SREs) with both enhancer and suppressor functions. Variants located in exonic regions can impact splicing through dysregulation of native splice sites, SREs, and cryptic splice site activation. While splicing dysregulation is considered primary disease-inducing mechanism of synonymous variants, its contribution toward disease phenotype of non-synonymous variants is underappreciated.
Methods: In this study, we analyzed 415 disease-causing and 120 neutral F9 exonic point variants including both synonymous and non-synonymous for their effect on splicing using a series of in silico splice site prediction tools, SRE prediction tools, and in vitro minigene assays.
Results: The use of splice site and SRE prediction tools in tandem provided better prediction but were not always in agreement with the minigene assays. The net effect of splicing dysregulation caused by variants was context dependent. Minigene assays revealed that perturbed splicing can be found.
Conclusion: Synonymous variants primarily cause disease phenotype via splicing dysregulation while additional mechanisms such as translation rate also play an important role. Splicing dysregulation is likely to contribute to the disease phenotype of several non-synonymous variants.
Keywords: hemophilia B; in silico splicing analysis; in vitro minigene assay; splicing dysregulation; synonymous variants.
© 2019 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
None declared.
Figures


Similar articles
-
Effects of 14 F9 synonymous codon variants on hemophilia B expression: Alteration of splicing along with protein expression.Hum Mutat. 2022 Jul;43(7):928-939. doi: 10.1002/humu.24377. Epub 2022 Apr 19. Hum Mutat. 2022. PMID: 35391506
-
Pleiotropic effects of different exonic nucleotide changes at the same position contribute to hemophilia B phenotypic variation.J Thromb Haemost. 2024 Apr;22(4):975-989. doi: 10.1016/j.jtha.2023.12.031. Epub 2024 Jan 4. J Thromb Haemost. 2024. PMID: 38184202
-
Apparent synonymous mutation F9 c.87A>G causes secretion failure by in-frame mutation with aberrant splicing.Thromb Res. 2019 Jul;179:95-103. doi: 10.1016/j.thromres.2019.04.022. Epub 2019 May 1. Thromb Res. 2019. PMID: 31102861
-
Code inside the codon: The role of synonymous mutations in regulating splicing machinery and its impact on disease.Mutat Res Rev Mutat Res. 2022 Jul-Dec;790:108444. doi: 10.1016/j.mrrev.2022.108444. Epub 2022 Oct 25. Mutat Res Rev Mutat Res. 2022. PMID: 36307006 Review.
-
The missing puzzle piece: splicing mutations.Int J Clin Exp Pathol. 2013 Nov 15;6(12):2675-82. eCollection 2013. Int J Clin Exp Pathol. 2013. PMID: 24294354 Free PMC article. Review.
Cited by
-
A Single Synonymous Variant (c.354G>A [p.P118P]) in ADAMTS13 Confers Enhanced Specific Activity.Int J Mol Sci. 2019 Nov 15;20(22):5734. doi: 10.3390/ijms20225734. Int J Mol Sci. 2019. PMID: 31731663 Free PMC article.
-
Effects of codon optimization on coagulation factor IX translation and structure: Implications for protein and gene therapies.Sci Rep. 2019 Oct 29;9(1):15449. doi: 10.1038/s41598-019-51984-2. Sci Rep. 2019. PMID: 31664102 Free PMC article.
-
In silico methods for predicting functional synonymous variants.Genome Biol. 2023 May 22;24(1):126. doi: 10.1186/s13059-023-02966-1. Genome Biol. 2023. PMID: 37217943 Free PMC article. Review.
-
A Code Within a Code: How Codons Fine-Tune Protein Folding in the Cell.Biochemistry (Mosc). 2021 Aug;86(8):976-991. doi: 10.1134/S0006297921080083. Biochemistry (Mosc). 2021. PMID: 34488574 Free PMC article. Review.
-
Hepatic lipase (LIPC) sequencing in individuals with extremely high and low high-density lipoprotein cholesterol levels.PLoS One. 2020 Dec 16;15(12):e0243919. doi: 10.1371/journal.pone.0243919. eCollection 2020. PLoS One. 2020. PMID: 33326441 Free PMC article.
References
-
- Balestra, D. , Scalet, D. , Pagani, F. , Rogalska, M. E. , Mari, R. , Bernardi, F. , & Pinotti, M. (2016). An exon‐specific U1snRNA induces a robust factor IX activity in mice expressing multiple human FIX splicing mutants. Molecular Therapy ‐ Nucleic Acids, 5(10), e370 10.1038/mtna.2016.77 - DOI - PMC - PubMed
-
- Bartoszewski, R. A. , Jablonsky, M. , Bartoszewska, S. , Stevenson, L. , Dai, Q. , Kappes, J. , … Bebok, Z. (2010). A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. Journal of Biological Chemistry, 285(37), 28741–28748. 10.1074/jbc.M110.154575 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

