Schlafen 3 knockout mice display gender-specific differences in weight gain, food efficiency, and expression of markers of intestinal epithelial differentiation, metabolism, and immune cell function
- PMID: 31260507
- PMCID: PMC6602453
- DOI: 10.1371/journal.pone.0219267
Schlafen 3 knockout mice display gender-specific differences in weight gain, food efficiency, and expression of markers of intestinal epithelial differentiation, metabolism, and immune cell function
Abstract
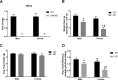
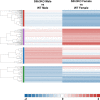
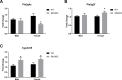
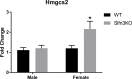
Self-renewal and differentiation are essential for intestinal epithelium absorptive functioning and adaptation to pathological states such as short gut syndrome, ulcers, and inflammatory bowel disease. The rodent Slfn3 and its human analog Slfn12 are critical in regulating intestinal epithelial differentiation. We sought to characterize intestinal function in Slfn3 knockout (KO) mice. Male and female pair-fed Slfn3KO mice gained less weight with decreased food efficiency than wild type (WT) mice, with more pronounced effects in females. RNA sequencing performed on intestinal mucosa of Slfn3KO and WT mice showed gene ontology decreases in cell adhesion molecule signaling, tumor necrosis factor receptor binding, and adaptive immune cell proliferation/functioning genes in Slfn3KO mice, with greater effects in females. qPCR analysis of fatty acid metabolism genes, Pla2g4c, Pla2g2f, and Cyp3c55 revealed an increase in Pla2g4c, and a decrease in Pla2g2f in Slfn3KO females. Additionally, adipogenesis genes, Fabp4 and Lpl were decreased and ketogenesis gene Hmgcs2 was increased in female Slfn3KO mice. Sequencing did not reveal significant changes in differentiation markers, so qPCR was utilized. Slfn3KO tended to have decreased expression of intestinal differentiation markers sucrase isomaltase, dipeptidyl peptidase 4, villin 1, and glucose transporter 1 (Glut1) vs. WT males, although these trends did not achieve statistical significance unless data from several markers was pooled. Differentiation markers, Glut2 and sodium-glucose transporter 1 (SGLT1), did show statistically significant sex-dependent differences. Glut2 mRNA was reduced in Slfn3KO females, while SGLT1 increased in Slfn3KO males. Notch2 and Cdx2 were only increased in female Slfn3KO mice. Although Slfn3KO mice gain less weight and decreased food efficiency, their biochemical phenotype is more subtle and suggests a complex interplay between gender effects, Slfn3, and another regulatory pathway yet to be identified that compensates for the chronic loss of Slfn3.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Vil-Cre specific Slfn3KO mice exhibit sex-specific differences in lung, stomach, cecum, kidney, and proximal colon differentiation markers and Slfn family members expression levels.Biochem Biophys Rep. 2023 Nov 23;36:101552. doi: 10.1016/j.bbrep.2023.101552. eCollection 2023 Dec. Biochem Biophys Rep. 2023. PMID: 38076659 Free PMC article.
-
Vil-Cre specific Schlafen 3 knockout mice exhibit sex-specific differences in intestinal differentiation markers and Schlafen family members expression levels.PLoS One. 2021 Oct 28;16(10):e0259195. doi: 10.1371/journal.pone.0259195. eCollection 2021. PLoS One. 2021. PMID: 34710177 Free PMC article.
-
Loss of Slfn3 induces a sex-dependent repair vulnerability after 50% bowel resection.Am J Physiol Gastrointest Liver Physiol. 2021 Jan 1;320(2):G136-G152. doi: 10.1152/ajpgi.00344.2020. Epub 2020 Nov 25. Am J Physiol Gastrointest Liver Physiol. 2021. PMID: 33237796 Free PMC article.
-
Wnt control of stem cells and differentiation in the intestinal epithelium.Exp Cell Res. 2005 Jun 10;306(2):357-63. doi: 10.1016/j.yexcr.2005.02.022. Epub 2005 Apr 7. Exp Cell Res. 2005. PMID: 15925592 Review.
-
Stem cells and the regulation of proliferation, differentiation and patterning in the intestinal epithelium: emerging insights from gene expression patterns, transgenic and gene ablation studies.Mech Dev. 2001 Mar;101(1-2):3-9. doi: 10.1016/s0925-4773(00)00557-8. Mech Dev. 2001. PMID: 11231054 Review.
Cited by
-
FABP4 in Paneth cells regulates antimicrobial protein expression to reprogram gut microbiota.Gut Microbes. 2022 Jan-Dec;14(1):2139978. doi: 10.1080/19490976.2022.2139978. Gut Microbes. 2022. PMID: 36519446 Free PMC article.
-
Vil-Cre specific Slfn3KO mice exhibit sex-specific differences in lung, stomach, cecum, kidney, and proximal colon differentiation markers and Slfn family members expression levels.Biochem Biophys Rep. 2023 Nov 23;36:101552. doi: 10.1016/j.bbrep.2023.101552. eCollection 2023 Dec. Biochem Biophys Rep. 2023. PMID: 38076659 Free PMC article.
-
Systematic review reveals sexually antagonistic knockouts in model organisms.Ecol Evol. 2022 Dec 28;12(12):e9671. doi: 10.1002/ece3.9671. eCollection 2022 Dec. Ecol Evol. 2022. PMID: 36619711 Free PMC article. Review.
-
Schlafen Family Intra-Regulation by IFN-α2 in Triple-Negative Breast Cancer.Cancers (Basel). 2023 Nov 30;15(23):5658. doi: 10.3390/cancers15235658. Cancers (Basel). 2023. PMID: 38067362 Free PMC article.
-
Vil-Cre specific Schlafen 3 knockout mice exhibit sex-specific differences in intestinal differentiation markers and Schlafen family members expression levels.PLoS One. 2021 Oct 28;16(10):e0259195. doi: 10.1371/journal.pone.0259195. eCollection 2021. PLoS One. 2021. PMID: 34710177 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

