Cloudy with a Chance of Insights: Context Dependent Gene Regulation and Implications for Evolutionary Studies
- PMID: 31261769
- PMCID: PMC6678813
- DOI: 10.3390/genes10070492
Cloudy with a Chance of Insights: Context Dependent Gene Regulation and Implications for Evolutionary Studies
Abstract
Research in various fields of evolutionary biology has shown that divergence in gene expression is a key driver for phenotypic evolution. An exceptional contribution of cis-regulatory divergence has been found to contribute to morphological diversification. In the light of these findings, the analysis of genome-wide expression data has become one of the central tools to link genotype and phenotype information on a more mechanistic level. However, in many studies, especially if general conclusions are drawn from such data, a key feature of gene regulation is often neglected. With our article, we want to raise awareness that gene regulation and thus gene expression is highly context dependent. Genes show tissue- and stage-specific expression. We argue that the regulatory context must be considered in comparative expression studies.
Keywords: Assay for Transposase-Accessible Chromatin using sequencing (ATACseq); ChIPseq; RNAseq; allele specific expression; chromatin; evolution; expression quantitative trait loci (eQTL); gene expression; gene regulation; genotype–phenotype map.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript.
Figures
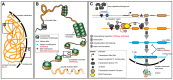
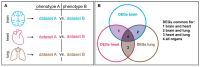
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

