Differences in Signaling Patterns on PI3K Inhibition Reveal Context Specificity in KRAS-Mutant Cancers
- PMID: 31262731
- PMCID: PMC6679718
- DOI: 10.1158/1535-7163.MCT-18-0727
Differences in Signaling Patterns on PI3K Inhibition Reveal Context Specificity in KRAS-Mutant Cancers
Abstract
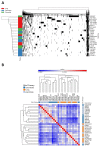
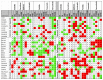
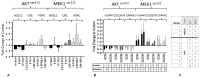
It is increasingly appreciated that drug response to different cancers driven by the same oncogene is different and may relate to differences in rewiring of signal transduction. We aimed to study differences in dynamic signaling changes within mutant KRAS (KRAS MT), non-small cell lung cancer (NSCLC), colorectal cancer, and pancreatic ductal adenocarcinoma (PDAC) cells. We used an antibody-based phosphoproteomic platform to study changes in 50 phosphoproteins caused by seven targeted anticancer drugs in a panel of 30 KRAS MT cell lines and cancer cells isolated from 10 patients with KRAS MT cancers. We report for the first time significant differences in dynamic signaling between colorectal cancer and NSCLC cell lines exposed to clinically relevant equimolar concentrations of the pan-PI3K inhibitor pictilisib including a lack of reduction of p-AKTser473 in colorectal cancer cell lines (P = 0.037) and lack of compensatory increase in p-MEK in NSCLC cell lines (P = 0.036). Differences in rewiring of signal transduction between tumor types driven by KRAS MT cancers exist and influence response to combination therapy using targeted agents.
©2019 American Association for Cancer Research.
Figures
References
-
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

