Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene loci
- PMID: 31262826
- PMCID: PMC6642405
- DOI: 10.1073/pnas.1903002116
Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene loci
Abstract
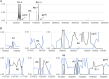
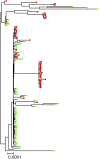
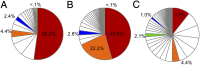
Global growth in antibiotic resistance is a major social problem. A high level of resistance to fluoroquinolones requires the concurrent presence of at least 3 mutations in the target proteins-2 in DNA gyrase (GyrA) and 1 in topoisomerase IV (ParC), which occur in a stepwise manner. In the Escherichia coli chromosome, the gyrA and parC loci are positioned about 1 Mb away from each other. Here we show that the 3 fluoroquinolone resistance mutations are tightly associated genetically in naturally occurring strains. In the latest pandemic uropathogenic and multidrug-resistant E. coli clonal group ST1193, the mutant variants of gyrA and parC were acquired not by a typical gradual, stepwise evolution but all at once. This happened as part of 11 simultaneous homologous recombination events involving 2 phylogenetically distant strains of E. coli, from an uropathogenic clonal complex ST14 and fluoroquinolone-resistant ST10. The gene exchanges swapped regions between 0.5 and 139 Kb in length (183 Kb total) spread along 976 Kb of chromosomal DNA around and between gyrA and parC loci. As a result, all 3 fluoroquinolone resistance mutations in GyrA and ParC have simultaneously appeared in ST1193. Based on molecular clock estimates, this potentially happened as recently as <12 y ago. Thus, naturally occurring homologous recombination events between 2 strains can involve numerous chromosomal gene locations simultaneously, resulting in the transfer of distant but tightly associated genetic mutations and emergence of a both highly pathogenic and antibiotic-resistant strain with a rapid global spread capability.
Keywords: Escherichia coli ST1193; QRDR mutations; homologous recombination; resistance to fluoroquinolones; urinary tract infections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Re: Pandemic Fluoroquinolone Resistant Escherichia coli Clone ST1193 Emerged via Simultaneous Homologous Recombinations in 11 Gene Loci.J Urol. 2020 Mar;203(3):466-467. doi: 10.1097/JU.0000000000000679.02. Epub 2019 Dec 3. J Urol. 2020. PMID: 31793814 No abstract available.
References
-
- Holmes A. H., et al. , Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187 (2016). - PubMed
-
- World Health Organization , Antimicrobial Resistance: Global Report on Surveillance 2014 (World Health Organization, 2014).
-
- Chen C. J., Huang Y. C., New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 20, 605–623 (2014). - PubMed
-
- Williamson D. A., Coombs G. W., Nimmo G. R., Staphylococcus aureus ‘Down Under’: Contemporary epidemiology of S. aureus in Australia, New Zealand, and the south west Pacific. Clin. Microbiol. Infect. 20, 597–604 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

