The Genome Sequence of the Anthelmintic-Susceptible New Zealand Haemonchus contortus
- PMID: 31263885
- PMCID: PMC6644846
- DOI: 10.1093/gbe/evz141
The Genome Sequence of the Anthelmintic-Susceptible New Zealand Haemonchus contortus
Abstract
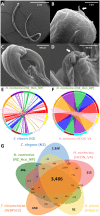
Internal parasitic nematodes are a global animal health issue causing drastic losses in livestock. Here, we report a H. contortus representative draft genome to serve as a genetic resource to the scientific community and support future experimental research of molecular mechanisms in related parasites. A de novo hybrid assembly was generated from PCR-free whole genome sequence data, resulting in a chromosome-level assembly that is 465 Mb in size encoding 22,341 genes. The genome sequence presented here is consistent with the genome architecture of the existing Haemonchus species and is a valuable resource for future studies regarding population genetic structures of parasitic nematodes. Additionally, comparative pan-genomics with other species of economically important parasitic nematodes have revealed highly open genomes and strong collinearities within the phylum Nematoda.
Keywords: Haemonchus contortus; anthelmintic-susceptible; genome; helminth; parasite.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Similar articles
-
Haemonchus contortus: Genome Structure, Organization and Comparative Genomics.Adv Parasitol. 2016;93:569-98. doi: 10.1016/bs.apar.2016.02.016. Epub 2016 Apr 11. Adv Parasitol. 2016. PMID: 27238013 Review.
-
The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery.Genome Biol. 2013 Aug 28;14(8):R88. doi: 10.1186/gb-2013-14-8-r88. Genome Biol. 2013. PMID: 23985316 Free PMC article.
-
The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus.Genome Biol. 2013 Aug 28;14(8):R89. doi: 10.1186/gb-2013-14-8-r89. Genome Biol. 2013. PMID: 23985341 Free PMC article.
-
Understanding Haemonchus contortus Better Through Genomics and Transcriptomics.Adv Parasitol. 2016;93:519-67. doi: 10.1016/bs.apar.2016.02.015. Epub 2016 Apr 5. Adv Parasitol. 2016. PMID: 27238012 Review.
-
Susceptible trichostrongyloid species mask presence of benzimidazole-resistant Haemonchus contortus in cattle.Parasit Vectors. 2021 Feb 8;14(1):101. doi: 10.1186/s13071-021-04593-w. Parasit Vectors. 2021. PMID: 33557939 Free PMC article.
Cited by
-
16S rRNA Gene Amplicon Profiling of the New Zealand Parasitic Blowfly Calliphora vicina.Microbiol Resour Announc. 2021 May 6;10(18):e00289-21. doi: 10.1128/MRA.00289-21. Microbiol Resour Announc. 2021. PMID: 33958401 Free PMC article.
-
Time-series transcriptomic profiling of larval exsheathment in a model parasitic nematode of veterinary importance.Front Cell Dev Biol. 2023 Nov 13;11:1257200. doi: 10.3389/fcell.2023.1257200. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38020897 Free PMC article. No abstract available.
-
Selection of Genome-Wide SNPs for Pooled Allelotyping Assays Useful for Population Monitoring.Genome Biol Evol. 2022 Mar 2;14(3):evac030. doi: 10.1093/gbe/evac030. Genome Biol Evol. 2022. PMID: 35179579 Free PMC article.
-
Multi-Omic Profiling, Structural Characterization, and Potent Inhibitor Screening of Evasion-Related Proteins of a Parasitic Nematode, Haemonchus contortus, Surviving Vaccine Treatment.Biomedicines. 2023 Jan 30;11(2):411. doi: 10.3390/biomedicines11020411. Biomedicines. 2023. PMID: 36830947 Free PMC article.
-
Characterization of the Complete Mitochondrial Genomes of Two Sibling Species of Parasitic Roundworms, Haemonchus contortus and Teladorsagia circumcincta.Front Genet. 2020 Oct 8;11:573395. doi: 10.3389/fgene.2020.573395. eCollection 2020. Front Genet. 2020. PMID: 33133162 Free PMC article.
References
-
- Bisset SA, Knight JS, Bouchet C.. 2014. A multiplex PCR-based method to identify strongylid parasite larvae recovered from ovine faecal cultures and/or pasture samples. Vet Parasitol. 200(1–2):117–127. - PubMed
-
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59.. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

