High CO2 Downregulates Skeletal Muscle Protein Anabolism via AMP-activated Protein Kinase α2-mediated Depressed Ribosomal Biogenesis
- PMID: 31264907
- PMCID: PMC6938128
- DOI: 10.1165/rcmb.2019-0061OC
High CO2 Downregulates Skeletal Muscle Protein Anabolism via AMP-activated Protein Kinase α2-mediated Depressed Ribosomal Biogenesis
Abstract
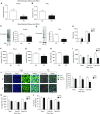
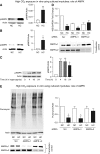
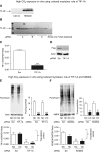
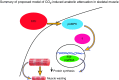
High CO2 retention, or hypercapnia, is associated with worse outcomes in patients with chronic pulmonary diseases. Skeletal muscle wasting is also an independent predictor of poor outcomes in patients with acute and chronic pulmonary diseases. Although previous evidence indicates that high CO2 accelerates skeletal muscle catabolism via AMPK (AMP-activated protein kinase)-FoxO3a-MuRF1 (E3-ubiquitin ligase muscle RING finger protein 1), little is known about the role of high CO2 in regulating skeletal muscle anabolism. In the present study, we investigated the potential role of high CO2 in attenuating skeletal muscle protein synthesis. We found that locomotor muscles from patients with chronic CO2 retention demonstrated depressed ribosomal gene expression in comparison with locomotor muscles from non-CO2-retaining individuals, and analysis of the muscle proteome of normo- and hypercapnic mice indicates reduction of important components of ribosomal structure and function. Indeed, mice chronically kept under a high-CO2 environment show evidence of skeletal muscle downregulation of ribosomal biogenesis and decreased protein synthesis as measured by the incorporation of puromycin into skeletal muscle. Hypercapnia did not regulate the mTOR pathway, and rapamycin-induced deactivation of mTOR did not cause a decrease in ribosomal gene expression. Loss-of-function studies in cultured myotubes showed that AMPKα2 regulates CO2-mediated reductions in ribosomal gene expression and protein synthesis. Although previous evidence has implicated TIF1A (transcription initiation factor-1α) and KDM2A (lysine-specific demethylase 2A) in AMPK-driven regulation of ribosomal gene expression, we found that these mediators were not required in the high CO2-induced depressed protein anabolism. Our research supports future studies targeting ribosomal biogenesis and protein synthesis to alleviate the effects of high CO2 on skeletal muscle turnover.
Keywords: anabolism; hypercapnia; ribosomal biogenesis; skeletal muscle.
Figures



Comment in
-
Antianabolic Effects of Hypercapnia: No Country for Strong Men.Am J Respir Cell Mol Biol. 2020 Jan;62(1):8-9. doi: 10.1165/rcmb.2019-0225ED. Am J Respir Cell Mol Biol. 2020. PMID: 31290693 Free PMC article. No abstract available.
References
-
- Marquis K, Debigaré R, Lacasse Y, LeBlanc P, Jobin J, Carrier G, et al. Midthigh muscle cross-sectional area is a better predictor of mortality than body mass index in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2002;166:809–813. - PubMed
-
- Shrikrishna D, Patel M, Tanner RJ, Seymour JM, Connolly BA, Puthucheary ZA, et al. Quadriceps wasting and physical inactivity in patients with COPD. Eur Respir J. 2012;40:1115–1122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

