Mapping of scaffold/matrix attachment regions in human genome: a data mining exercise
- PMID: 31265077
- PMCID: PMC6698742
- DOI: 10.1093/nar/gkz562
Mapping of scaffold/matrix attachment regions in human genome: a data mining exercise
Abstract
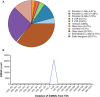
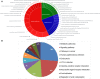
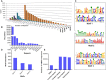
Scaffold/matrix attachment regions (S/MARs) are DNA elements that serve to compartmentalize the chromatin into structural and functional domains. These elements are involved in control of gene expression which governs the phenotype and also plays role in disease biology. Therefore, genome-wide understanding of these elements holds great therapeutic promise. Several attempts have been made toward identification of S/MARs in genomes of various organisms including human. However, a comprehensive genome-wide map of human S/MARs is yet not available. Toward this objective, ChIP-Seq data of 14 S/MAR binding proteins were analyzed and the binding site coordinates of these proteins were used to prepare a non-redundant S/MAR dataset of human genome. Along with co-ordinate (location) details of S/MARs, the dataset also revealed details of S/MAR features, namely, length, inter-SMAR length (the chromatin loop size), nucleotide repeats, motif abundance, chromosomal distribution and genomic context. S/MARs identified in present study and their subsequent analysis also suggests that these elements act as hotspots for integration of retroviruses. Therefore, these data will help toward better understanding of genome functioning and designing effective anti-viral therapeutics. In order to facilitate user friendly browsing and retrieval of the data obtained in present study, a web interface, MARome (http://bioinfo.net.in/MARome), has been developed.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Chromatin loops are selectively anchored using scaffold/matrix-attachment regions.J Cell Sci. 2004 Mar 1;117(Pt 7):999-1008. doi: 10.1242/jcs.00976. J Cell Sci. 2004. PMID: 14996931
-
Correlations between scaffold/matrix attachment region (S/MAR) binding activity and DNA duplex destabilization energy.J Mol Biol. 2006 Apr 28;358(2):597-613. doi: 10.1016/j.jmb.2005.11.073. Epub 2005 Dec 9. J Mol Biol. 2006. PMID: 16516920
-
Identification and mapping of nuclear matrix-attachment regions in a one megabase locus of human chromosome 19q13.12: long-range correlation of S/MARs and gene positions.J Cell Biochem. 2002;84(3):590-600. J Cell Biochem. 2002. PMID: 11813264
-
MARs and MARBPs: key modulators of gene regulation and disease manifestation.Subcell Biochem. 2007;41:213-30. Subcell Biochem. 2007. PMID: 17484130 Review.
-
Scaffold/Matrix Attachment Regions and intrinsic DNA curvature.Biochemistry (Mosc). 2006 May;71(5):481-8. doi: 10.1134/s0006297906050038. Biochemistry (Mosc). 2006. PMID: 16732725 Review.
Cited by
-
Species-Specific Differences in Sperm Chromatin Decondensation Between Eutherian Mammals Underlie Distinct Lysis Requirements.Front Cell Dev Biol. 2021 Apr 30;9:669182. doi: 10.3389/fcell.2021.669182. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33996825 Free PMC article.
-
Improved Functionality of Integration-Deficient Lentiviral Vectors (IDLVs) by the Inclusion of IS2 Protein Docks.Pharmaceutics. 2021 Aug 6;13(8):1217. doi: 10.3390/pharmaceutics13081217. Pharmaceutics. 2021. PMID: 34452178 Free PMC article.
-
Feasibility of machine learning-based modeling and prediction to assess osteosarcoma outcomes.Sci Rep. 2025 May 19;15(1):17386. doi: 10.1038/s41598-025-00179-z. Sci Rep. 2025. PMID: 40389469 Free PMC article.
-
Purine-Rich Element Binding Protein Alpha, a Nuclear Matrix Protein, Has a Role in Prostate Cancer Progression.Int J Mol Sci. 2024 Jun 24;25(13):6911. doi: 10.3390/ijms25136911. Int J Mol Sci. 2024. PMID: 39000020 Free PMC article. Review.
-
Nuclear architecture and the structural basis of mitotic memory.Chromosome Res. 2023 Feb 2;31(1):8. doi: 10.1007/s10577-023-09714-y. Chromosome Res. 2023. PMID: 36725757 Review.
References
-
- Heng H.H.Q. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J. Cell Sci. 2004; 117:999–1008. - PubMed
-
- Capco D.G., Wan K.M., Penman S., Weber K., Franke W.W., Fyne C.-T.. The nuclear matrix: three-dimensional architecture and protein composition. Cell. 1982; 29:847–858. - PubMed
-
- Razin S. V, Gromova I.I., Iarovaia O. V. Specificity and functional significance of DNA interaction with the nuclear matrix: new approaches to clarify the old questions. Int. Rev. Cytol. 1995; 162B:405–448. - PubMed
-
- Stein G.S., Zaidi S.K., Braastad C.D., Montecino M., van Wijnen A.J., Choi J.-Y., Stein J.L., Lian J.B., Javed A.. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol. 2003; 13:584–592. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

