Overexpression of an LaeA-like Methyltransferase Upregulates Secondary Metabolite Production in Aspergillus nidulans
- PMID: 31265232
- PMCID: PMC7310610
- DOI: 10.1021/acschembio.9b00380
Overexpression of an LaeA-like Methyltransferase Upregulates Secondary Metabolite Production in Aspergillus nidulans
Abstract
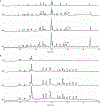
Fungal secondary metabolites (SMs) include medically valuable compounds as well as compounds that are toxic, carcinogenic, and/or contributors to fungal pathogenesis. It is consequently important to understand the regulation of fungal secondary metabolism. McrA is a recently discovered transcription factor that negatively regulates fungal secondary metabolism. Deletion of mcrA (mcrAΔ), the gene encoding McrA, results in upregulation of many SMs and alters the expression of more than 1000 genes. One gene strongly upregulated by the deletion of mcrA is llmG, a putative methyl transferase related to LaeA, a major regulator of secondary metabolism. We artificially upregulated llmG by replacing its promoter with strong constitutive promoters in strains carrying either wild-type mcrA or mcrAΔ. Upregulation of llmG on various media resulted in increased production of the important toxin sterigmatocystin and compounds from at least six major SM pathways. llmG is, thus, a master SM regulator. mcrAΔ generally resulted in greater upregulation of SMs than upregulation of llmG, indicating that the full effects of mcrA on secondary metabolism involve genes in addition to llmG. However, the combination of mcrAΔ and upregulation of llmG generally resulted in greater compound production than mcrAΔ alone (in one case more than 460 times greater than the control). This result indicates that deletion of mcrA and/or upregulation of llmG can likely be combined with other strategies for eliciting SM production to greater levels than can be obtained with any single strategy.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
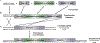


References
-
- Martín JF (1998) New Aspects of genes and enzymes for β-lactam antibiotic biosynthesis. Appl. Microbiol. Biotechnol. Heidelb 50 (1), 1–15. - PubMed
-
- Keller NP, Turner G, Bennett JW (2005) Fungal secondary metabolism-from biochemistry to genomics. Nat. Rev. Microbiol 3 (12), 937–947. - PubMed
-
- Peláez F. Biological Activities of Fungal Metabolites. https://www.taylorfrancis.com/
-
- Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP (2006) Genomic mining for Aspergillus natural products. Chem. Biol 13 (1), 31–37. - PubMed
-
- Hoffmeister D, Keller NP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep 24 (2), 393–416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

