Acylpeptide hydrolase is a novel regulator of KRAS plasma membrane localization and function
- PMID: 31266814
- PMCID: PMC6703705
- DOI: 10.1242/jcs.232132
Acylpeptide hydrolase is a novel regulator of KRAS plasma membrane localization and function
Abstract
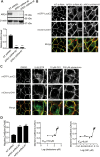
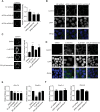
The primary site for KRAS signaling is the inner leaflet of the plasma membrane (PM). We previously reported that oxanthroquinone G01 (G01) inhibited KRAS PM localization and blocked KRAS signaling. In this study, we identified acylpeptide hydrolase (APEH) as a molecular target of G01. APEH formed a stable complex with biotinylated G01, and the enzymatic activity of APEH was inhibited by G01. APEH knockdown caused profound mislocalization of KRAS and reduced clustering of KRAS that remained PM localized. APEH knockdown also disrupted the PM localization of phosphatidylserine (PtdSer), a lipid critical for KRAS PM binding and clustering. The mislocalization of KRAS was fully rescued by ectopic expression of APEH in knockdown cells. APEH knockdown disrupted the endocytic recycling of epidermal growth factor receptor and transferrin receptor, suggesting that abrogation of recycling endosome function was mechanistically linked to the loss of KRAS and PtdSer from the PM. APEH knockdown abrogated RAS-RAF-MAPK signaling in cells expressing the constitutively active (oncogenic) mutant of KRAS (KRASG12V), and selectively inhibited the proliferation of KRAS-transformed pancreatic cancer cells. Taken together, these results identify APEH as a novel drug target for a potential anti-KRAS therapeutic.
Keywords: Acylpeptide hydrolase; KRAS; Oxanthroquinone; Plasma membrane; Recycling endosome; Sphingomyelin metabolism.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures






Similar articles
-
An oxanthroquinone derivative that disrupts RAS plasma membrane localization inhibits cancer cell growth.J Biol Chem. 2018 Aug 31;293(35):13696-13706. doi: 10.1074/jbc.RA118.003907. Epub 2018 Jul 3. J Biol Chem. 2018. PMID: 29970615 Free PMC article.
-
Components of the phosphatidylserine endoplasmic reticulum to plasma membrane transport mechanism as targets for KRAS inhibition in pancreatic cancer.Proc Natl Acad Sci U S A. 2021 Dec 21;118(51):e2114126118. doi: 10.1073/pnas.2114126118. Proc Natl Acad Sci U S A. 2021. PMID: 34903667 Free PMC article.
-
Targeting plasma membrane phosphatidylserine content to inhibit oncogenic KRAS function.Life Sci Alliance. 2019 Aug 26;2(5):e201900431. doi: 10.26508/lsa.201900431. Print 2019 Oct. Life Sci Alliance. 2019. PMID: 31451509 Free PMC article.
-
Inhibitors of K-Ras plasma membrane localization.Enzymes. 2013;33 Pt A:249-65. doi: 10.1016/B978-0-12-416749-0.00011-7. Epub 2013 Aug 8. Enzymes. 2013. PMID: 25033808 Review.
-
Is Nanoclustering essential for all oncogenic KRas pathways? Can it explain why wild-type KRas can inhibit its oncogenic variant?Semin Cancer Biol. 2019 Feb;54:114-120. doi: 10.1016/j.semcancer.2018.01.002. Epub 2018 Jan 5. Semin Cancer Biol. 2019. PMID: 29307569 Review.
Cited by
-
RAS Function in cancer cells: translating membrane biology and biochemistry into new therapeutics.Biochem J. 2020 Aug 14;477(15):2893-2919. doi: 10.1042/BCJ20190839. Biochem J. 2020. PMID: 32797215 Free PMC article. Review.
-
MTMR regulates KRAS function by controlling plasma membrane levels of phospholipids.J Cell Biol. 2025 Jul 7;224(7):e202403126. doi: 10.1083/jcb.202403126. Epub 2025 May 2. J Cell Biol. 2025. PMID: 40314454
-
Cryo-EM structure of acylpeptide hydrolase reveals substrate selection by multimerization and a multi-state serine-protease triad.Chem Sci. 2022 May 18;13(24):7132-7142. doi: 10.1039/d2sc02276a. eCollection 2022 Jun 22. Chem Sci. 2022. PMID: 35799812 Free PMC article.
-
Precise and efficient silencing of mutant KrasG12D by CRISPR-CasRx controls pancreatic cancer progression.Theranostics. 2020 Sep 16;10(25):11507-11519. doi: 10.7150/thno.46642. eCollection 2020. Theranostics. 2020. PMID: 33052229 Free PMC article.
-
Differential Distribution and Activity Profile of Acylpeptide Hydrolase in the Rat Seminiferous Epithelium.Biomedicines. 2022 Jul 4;10(7):1591. doi: 10.3390/biomedicines10071591. Biomedicines. 2022. PMID: 35884896 Free PMC article.
References
-
- Bergamo P., Palmieri G., Cocca E., Ferrandino I., Gogliettino M., Monaco A., Maurano F. and Rossi M. (2016). Adaptive response activated by dietary cis9, trans11 conjugated linoleic acid prevents distinct signs of gliadin-induced enteropathy in mice. Eur. J. Nutr. 55, 729-740. 10.1007/s00394-015-0893-2 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

