New Insights into the Intrinsic and Extrinsic Factors That Shape the Human Skin Microbiome
- PMID: 31266865
- PMCID: PMC6606800
- DOI: 10.1128/mBio.00839-19
New Insights into the Intrinsic and Extrinsic Factors That Shape the Human Skin Microbiome
Abstract
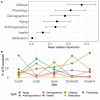
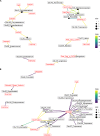
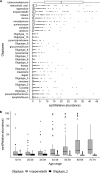
Despite recognition that biogeography and individuality shape the function and composition of the human skin microbiome, we know little about how extrinsic and intrinsic host factors influence its composition. To explore the contributions of these factors to skin microbiome variation, we profiled the bacterial microbiomes of 495 North American subjects (ages, 9 to 78 years) at four skin surfaces plus the oral epithelium using 16S rRNA gene amplicon sequencing. We collected subject metadata, including host physiological parameters, through standardized questionnaires and noninvasive biophysical methods. Using a combination of statistical modeling tools, we found that demographic, lifestyle, and physiological factors collectively explained 12 to 20% of the variability in microbiome composition. The influence of health factors was strongest on the oral microbiome. Associations between host factors and the skin microbiome were generally dominated by operational taxonomic units (OTUs) affiliated with the Clostridiales and Prevotella A subset of the correlations between microbial features and host attributes were site specific. To further explore the relationship between age and the skin microbiome of the forehead, we trained a Random Forest regression model to predict chronological age from microbial features. Age was associated mostly with two mutually coexcluding Corynebacterium OTUs. Furthermore, skin aging variables (wrinkles and hyperpigmented spots) were independently correlated to these taxa.IMPORTANCE Many studies have highlighted the importance of body site and individuality in shaping the composition of the human skin microbiome, but we still have a poor understanding of how extrinsic (e.g., lifestyle) and intrinsic (e.g., age) factors influence its composition. We characterized the bacterial microbiomes of North American volunteers at four skin sites and the mouth. We also collected extensive subject metadata and measured several host physiological parameters. Integration of host and microbial features showed that the skin microbiome was predominantly associated with demographic, lifestyle, and physiological factors. Furthermore, we uncovered reproducible associations between chronological age, skin aging, and members of the genus Corynebacterium Our work provides new understanding of the role of host selection and lifestyle in shaping skin microbiome composition. It also contributes to a more comprehensive appreciation of the factors that drive interindividual skin microbiome variation.
Keywords: Corynebacterium; age; demographic; forehead; host lifestyle; host physiology; metadata; scalp; skin microbiome.
Copyright © 2019 Dimitriu et al.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

