mRNA Degradation Rates Are Coupled to Metabolic Status in Mycobacterium smegmatis
- PMID: 31266866
- PMCID: PMC6606801
- DOI: 10.1128/mBio.00957-19
mRNA Degradation Rates Are Coupled to Metabolic Status in Mycobacterium smegmatis
Abstract
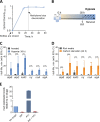
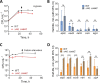
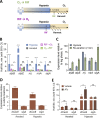
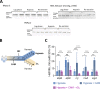
The success of Mycobacterium tuberculosis as a human pathogen is due in part to its ability to survive stress conditions, such as hypoxia or nutrient deprivation, by entering nongrowing states. In these low-metabolism states, M. tuberculosis can tolerate antibiotics and develop genetically encoded antibiotic resistance, making its metabolic adaptation to stress crucial for survival. Numerous bacteria, including M. tuberculosis, have been shown to reduce their rates of mRNA degradation under growth limitation and stress. While the existence of this response appears to be conserved across species, the underlying bacterial mRNA stabilization mechanisms remain unknown. To better understand the biology of nongrowing mycobacteria, we sought to identify the mechanistic basis of mRNA stabilization in the nonpathogenic model Mycobacterium smegmatis We found that mRNA half-life was responsive to energy stress, with carbon starvation and hypoxia causing global mRNA stabilization. This global stabilization was rapidly reversed when hypoxia-adapted cultures were reexposed to oxygen, even in the absence of new transcription. The stringent response and RNase levels did not explain mRNA stabilization, nor did transcript abundance. This led us to hypothesize that metabolic changes during growth cessation impact the activities of degradation proteins, increasing mRNA stability. Indeed, bedaquiline and isoniazid, two drugs with opposing effects on cellular energy status, had opposite effects on mRNA half-lives in growth-arrested cells. Taken together, our results indicate that mRNA stability in mycobacteria is not directly regulated by growth status but rather is dependent on the status of energy metabolism.IMPORTANCE The logistics of tuberculosis therapy are difficult, requiring multiple drugs for many months. Mycobacterium tuberculosis survives in part by entering nongrowing states in which it is metabolically less active and thus less susceptible to antibiotics. Basic knowledge on how M. tuberculosis survives during these low-metabolism states is incomplete, and we hypothesize that optimized energy resource management is important. Here, we report that slowed mRNA turnover is a common feature of mycobacteria under energy stress but is not dependent on the mechanisms that have generally been postulated in the literature. Finally, we found that mRNA stability and growth status can be decoupled by a drug that causes growth arrest but increases metabolic activity, indicating that mRNA stability responds to metabolic status rather than to growth rate per se Our findings suggest a need to reorient studies of global mRNA stabilization to identify novel mechanisms that are presumably responsible.
Keywords: ATP; Mycobacterium smegmatis; Mycobacterium tuberculosis; carbon starvation; hypoxia; mRNA degradation; mRNA stability; stress response; tuberculosis.
Copyright © 2019 Vargas-Blanco et al.
Figures
References
-
- Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, Lee EG, Park SK, Raleigh JA, Cho SN, McMurray DN, Flynn JL, Barry CE III.. 2008. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun 76:2333–2340. doi:10.1128/IAI.01515-07. - DOI - PMC - PubMed
-
- Belton M, Brilha S, Manavaki R, Mauri F, Nijran K, Hong YT, Patel NH, Dembek M, Tezera L, Green J, Moores R, Aigbirhio F, Al-Nahhas A, Fryer TD, Elkington PT, Friedland JS. 2016. Hypoxia and tissue destruction in pulmonary TB. Thorax 71:1145–1153. doi:10.1136/thoraxjnl-2015-207402. - DOI - PMC - PubMed
-
- Albertson MH, Nyström T, Kjelleberg S. 1990. Functional mRNA half-lives in the marine Vibrio sp. S14 during starvation and recovery. Microbiology 136:2195–2199. doi:10.1099/00221287-136-11-2195. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
