RadD Contributes to R-Loop Avoidance in Sub-MIC Tobramycin
- PMID: 31266870
- PMCID: PMC6606805
- DOI: 10.1128/mBio.01173-19
RadD Contributes to R-Loop Avoidance in Sub-MIC Tobramycin
Abstract
We have previously identified Vibrio cholerae mutants in which the stress response to subinhibitory concentrations of aminoglycoside is altered. One gene identified, VC1636, encodes a putative DNA/RNA helicase, recently named RadD in Escherichia coli Here we combined extensive genetic characterization and high-throughput approaches in order to identify partners and molecular mechanisms involving RadD. We show that double-strand DNA breaks (DSBs) are formed upon subinhibitory tobramycin treatment in the absence of radD and recBCD and that formation of these DSBs can be overcome by RNase H1 overexpression. Loss of RNase H1, or of the transcription-translation coupling factor EF-P, is lethal in the radD deletion mutant. We propose that R-loops are formed upon sublethal aminoglycoside treatment, leading to the formation of DSBs that can be repaired by the RecBCD homologous recombination pathway, and that RadD counteracts such R-loop accumulation. We discuss how R-loops that can occur upon translation-transcription uncoupling could be the link between tobramycin treatment and DNA break formation.IMPORTANCE Bacteria frequently encounter low concentrations of antibiotics. Active antibiotics are commonly detected in soil and water at concentrations much below lethal concentration. Although sub-MICs of antibiotics do not kill bacteria, they can have a major impact on bacterial populations by contributing to the development of antibiotic resistance through mutations in originally sensitive bacteria or acquisition of DNA from resistant bacteria. It was shown that concentrations as low as 100-fold below the MIC can actually lead to the selection of antibiotic-resistant cells. We seek to understand how bacterial cells react to such antibiotic concentrations using E. coli, the Gram-negative bacterial paradigm, and V. cholerae, the causative agent of cholera. Our findings shed light on the processes triggered at the DNA level by antibiotics targeting translation, how damage occurs, and what the bacterial strategies are to respond to such DNA damage.
Keywords: DNA repair; R-loop; antibiotic resistance.
Copyright © 2019 Negro et al.
Figures



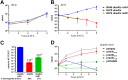

Similar articles
-
Identification of genes involved in low aminoglycoside-induced SOS response in Vibrio cholerae: a role for transcription stalling and Mfd helicase.Nucleic Acids Res. 2014 Feb;42(4):2366-79. doi: 10.1093/nar/gkt1259. Epub 2013 Dec 6. Nucleic Acids Res. 2014. PMID: 24319148 Free PMC article.
-
Escherichia coli RadD Protein Functionally Interacts with the Single-stranded DNA-binding Protein.J Biol Chem. 2016 Sep 23;291(39):20779-86. doi: 10.1074/jbc.M116.736223. Epub 2016 Aug 12. J Biol Chem. 2016. PMID: 27519413 Free PMC article.
-
RpoS plays a central role in the SOS induction by sub-lethal aminoglycoside concentrations in Vibrio cholerae.PLoS Genet. 2013;9(4):e1003421. doi: 10.1371/journal.pgen.1003421. Epub 2013 Apr 11. PLoS Genet. 2013. PMID: 23613664 Free PMC article.
-
DksA and DNA double-strand break repair.Curr Genet. 2019 Dec;65(6):1297-1300. doi: 10.1007/s00294-019-00983-x. Epub 2019 May 10. Curr Genet. 2019. PMID: 31076845 Review.
-
R-LOOPs on Short Tandem Repeat Expansion Disorders in Neurodegenerative Diseases.Mol Neurobiol. 2023 Dec;60(12):7185-7195. doi: 10.1007/s12035-023-03531-4. Epub 2023 Aug 4. Mol Neurobiol. 2023. PMID: 37540313 Review.
Cited by
-
RNA polymerase stalling-derived genome instability underlies ribosomal antibiotic efficacy and resistance evolution.Nat Commun. 2024 Aug 3;15(1):6579. doi: 10.1038/s41467-024-50917-6. Nat Commun. 2024. PMID: 39097616 Free PMC article.
-
Aminoglycoside tolerance in Vibrio cholerae engages translational reprogramming associated with queuosine tRNA modification.Elife. 2025 Jan 6;13:RP96317. doi: 10.7554/eLife.96317. Elife. 2025. PMID: 39761105 Free PMC article.
-
Sleeping ribosomes: Bacterial signaling triggers RaiA mediated persistence to aminoglycosides.iScience. 2021 Sep 14;24(10):103128. doi: 10.1016/j.isci.2021.103128. eCollection 2021 Oct 22. iScience. 2021. PMID: 34611612 Free PMC article.
-
Issues beyond resistance: inadequate antibiotic therapy and bacterial hypervirulence.FEMS Microbes. 2020 Oct 17;1(1):xtaa004. doi: 10.1093/femsmc/xtaa004. eCollection 2020 Sep. FEMS Microbes. 2020. PMID: 37333955 Free PMC article. Review.
-
Resolving Toxic DNA repair intermediates in every E. coli replication cycle: critical roles for RecG, Uup and RadD.Nucleic Acids Res. 2020 Sep 4;48(15):8445-8460. doi: 10.1093/nar/gkaa579. Nucleic Acids Res. 2020. PMID: 32644157 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
