An EDS1-SAG101 Complex Is Essential for TNL-Mediated Immunity in Nicotiana benthamiana
- PMID: 31266900
- PMCID: PMC6790086
- DOI: 10.1105/tpc.19.00099
An EDS1-SAG101 Complex Is Essential for TNL-Mediated Immunity in Nicotiana benthamiana
Abstract
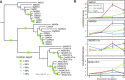
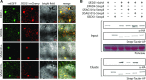
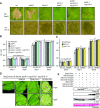
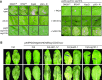
Heterodimeric complexes containing the lipase-like protein ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) are regarded as central regulators of plant innate immunity. In this context, a complex of EDS1 with PHYTOALEXIN DEFICIENT4 (PAD4) is required for basal resistance and signaling downstream of immune receptors containing an N-terminal Toll-interleukin-1 receptor-like domain (TNLs) in Arabidopsis (Arabidopsis thaliana). Here we analyze EDS1 functions in the model Solanaceous plant Nicotiana benthamiana (Nb). Stable Nb mutants deficient in EDS1 complexes are not impaired in basal resistance, a finding which contradicts a general role for EDS1 in immunity. In Nb, PAD4 demonstrated no detectable immune functions, but TNL-mediated resistance responses required EDS1 complexes incorporating a SENESCENCE ASSOCIATED GENE101 (SAG101) isoform. Intriguingly, SAG101 is restricted to those genomes also encoding TNL receptors, and we propose it may be required for TNL-mediated immune signaling in most plants, except the Brassicaceae. Transient complementation in Nb was used for accelerated mutational analyses while avoiding complex biotic interactions. We identify a large surface essential for EDS1-SAG101 immune functions that extends from the N-terminal lipase domains to the C-terminal EDS1-PAD4 domains and might mediate interaction partner recruitment. Furthermore, this work demonstrates the value of genetic resources in Nb, which will facilitate elucidation of EDS1 functions.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures




Comment in
-
Die Another Way: An EDS1-SAG101 Complex Mediates TNL Immunity in Solanaceous Plants.Plant Cell. 2019 Oct;31(10):2289-2290. doi: 10.1105/tpc.19.00570. Epub 2019 Jul 29. Plant Cell. 2019. PMID: 31358648 Free PMC article. No abstract available.
Similar articles
-
A Coevolved EDS1-SAG101-NRG1 Module Mediates Cell Death Signaling by TIR-Domain Immune Receptors.Plant Cell. 2019 Oct;31(10):2430-2455. doi: 10.1105/tpc.19.00118. Epub 2019 Jul 16. Plant Cell. 2019. PMID: 31311833 Free PMC article.
-
Pathogen effector recognition-dependent association of NRG1 with EDS1 and SAG101 in TNL receptor immunity.Nat Commun. 2021 Jun 7;12(1):3335. doi: 10.1038/s41467-021-23614-x. Nat Commun. 2021. PMID: 34099661 Free PMC article.
-
EDS1 complexes are not required for PRR responses and execute TNL-ETI from the nucleus in Nicotiana benthamiana.New Phytol. 2022 Dec;236(6):2249-2264. doi: 10.1111/nph.18511. Epub 2022 Oct 12. New Phytol. 2022. PMID: 36151929
-
EDS1 signalling: At the nexus of intracellular and surface receptor immunity.Curr Opin Plant Biol. 2021 Aug;62:102039. doi: 10.1016/j.pbi.2021.102039. Epub 2021 Apr 27. Curr Opin Plant Biol. 2021. PMID: 33930849 Review.
-
Plant immunity: the EDS1 regulatory node.Curr Opin Plant Biol. 2005 Aug;8(4):383-9. doi: 10.1016/j.pbi.2005.05.010. Curr Opin Plant Biol. 2005. PMID: 15939664 Review.
Cited by
-
Plant NLR immunity activation and execution: a biochemical perspective.Open Biol. 2024 Jan;14(1):230387. doi: 10.1098/rsob.230387. Epub 2024 Jan 24. Open Biol. 2024. PMID: 38262605 Free PMC article. Review.
-
Molecular insights into the biochemical functions and signalling mechanisms of plant NLRs.Mol Plant Pathol. 2022 Jun;23(6):772-780. doi: 10.1111/mpp.13195. Epub 2022 Mar 30. Mol Plant Pathol. 2022. PMID: 35355394 Free PMC article. Review.
-
Transcription Factor Pso9TF Assists Xinjiang Wild Myrobalan Plum (Prunus sogdiana) PsoRPM3 Disease Resistance Protein to Resist Meloidogyne incognita.Plants (Basel). 2021 Jul 29;10(8):1561. doi: 10.3390/plants10081561. Plants (Basel). 2021. PMID: 34451606 Free PMC article.
-
Sensor NLR immune proteins activate oligomerization of their NRC helpers in response to plant pathogens.EMBO J. 2023 Mar 1;42(5):e111519. doi: 10.15252/embj.2022111519. Epub 2022 Dec 29. EMBO J. 2023. PMID: 36579501 Free PMC article.
-
An Overview of PRR- and NLR-Mediated Immunities: Conserved Signaling Components across the Plant Kingdom That Communicate Both Pathways.Int J Mol Sci. 2022 Oct 26;23(21):12974. doi: 10.3390/ijms232112974. Int J Mol Sci. 2022. PMID: 36361764 Free PMC article. Review.
References
-
- Adlung N., Bonas U. (2017). Dissecting virulence function from recognition: Cell death suppression in Nicotiana benthamiana by XopQ/HopQ1-family effectors relies on EDS1-dependent immunity. Plant J. 91: 430–442. - PubMed
-
- Adlung N., Prochaska H., Thieme S., Banik A., Blüher D., John P., Nagel O., Schulze S., Gantner J., Delker C., Stuttmann J., Bonas U. (2016). Non-host resistance induced by the Xanthomonas effector XopQ is widespread within the genus Nicotiana and functionally depends on EDS1. Front. Plant Sci. 7: 1796. - PMC - PubMed
-
- Bent A.F., Kunkel B.N., Dahlbeck D., Brown K.L., Schmidt R., Giraudat J., Leung J., Staskawicz B.J. (1994). RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

