Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice
- PMID: 31266936
- PMCID: PMC6606613
- DOI: 10.1038/s41467-019-10366-y
Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice
Abstract
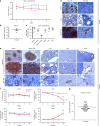
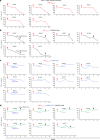
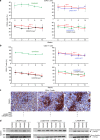
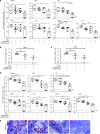
Elimination of HIV-1 requires clearance and removal of integrated proviral DNA from infected cells and tissues. Here, sequential long-acting slow-effective release antiviral therapy (LASER ART) and CRISPR-Cas9 demonstrate viral clearance in latent infectious reservoirs in HIV-1 infected humanized mice. HIV-1 subgenomic DNA fragments, spanning the long terminal repeats and the Gag gene, are excised in vivo, resulting in elimination of integrated proviral DNA; virus is not detected in blood, lymphoid tissue, bone marrow and brain by nested and digital-droplet PCR as well as RNAscope tests. No CRISPR-Cas9 mediated off-target effects are detected. Adoptive transfer of human immunocytes from dual treated, virus-free animals to uninfected humanized mice fails to produce infectious progeny virus. In contrast, HIV-1 is readily detected following sole LASER ART or CRISPR-Cas9 treatment. These data provide proof-of-concept that permanent viral elimination is possible.
Conflict of interest statement
K.K. and RK are named inventors on patents that cover the viral gene editing technology that is the subject of this article. HEG and BE are named inventors on patents that cover the medicinal and polymer chemistry technologies that were also employed in this manuscript. In addition to the foregoing interests, KK is a co-founder, board member, scientific advisor, and holds equity in Excision Biotherapeutics, a biotech start-up that has licensed the viral gene editing technology from Temple University for commercial development and clinical trials. H.E.G. is the founder of Brain First, a second independent biotech start-up and is the operative head of the Nebraska Nanomedicine Production Plant, a good manufacturing program facility that was responsible for synthesizing the long-acting slow effective release antiretroviral therapies used in the accompanying report. The authors declare that this work was produced solely by the authors and that no other individuals or entities influenced any aspects of the work including, but not limited to, the study conception and design; data acquisition, analysis and interpretation; and writing of the manuscript. No other entities provided funds for the work. The authors further declare that they have received no financial compensation from any other third parties for any aspects of the published work. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS036126/NS/NINDS NIH HHS/United States
- P01 DA028555/DA/NIDA NIH HHS/United States
- P30 MH062261/MH/NIMH NIH HHS/United States
- R01 DA042706/DA/NIDA NIH HHS/United States
- P01 DA037830/DA/NIDA NIH HHS/United States
- R01 NS087971/NS/NINDS NIH HHS/United States
- R01 MH104147/MH/NIMH NIH HHS/United States
- R01 DA013137/DA/NIDA NIH HHS/United States
- P30 AI078498/AI/NIAID NIH HHS/United States
- R24 OD018546/OD/NIH HHS/United States
- R01 MH115860/MH/NIMH NIH HHS/United States
- P01 NS043985/NS/NINDS NIH HHS/United States
- P30 MH092177/MH/NIMH NIH HHS/United States
- R01 AG043540/AG/NIA NIH HHS/United States
- R01 MH110360/MH/NIMH NIH HHS/United States
- R01 NS034239/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

