Enhancer accessibility and CTCF occupancy underlie asymmetric TAD architecture and cell type specific genome topology
- PMID: 31266948
- PMCID: PMC6606583
- DOI: 10.1038/s41467-019-10725-9
Enhancer accessibility and CTCF occupancy underlie asymmetric TAD architecture and cell type specific genome topology
Abstract
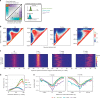
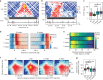
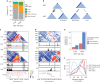
Cohesin and CTCF are master regulators of genome topology. How these ubiquitous proteins contribute to cell-type specific genome structure is poorly understood. Here, we explore quantitative aspects of topologically associated domains (TAD) between pluripotent embryonic stem cells (ESC) and lineage-committed cells. ESCs exhibit permissive topological configurations which manifest themselves as increased inter- TAD interactions, weaker intra-TAD interactions, and a unique intra-TAD connectivity whereby one border makes pervasive interactions throughout the domain. Such 'stripe' domains are associated with both poised and active chromatin landscapes and transcription is not a key determinant of their structure. By tracking the developmental dynamics of stripe domains, we show that stripe formation is linked to the functional state of the cell through cohesin loading at lineage-specific enhancers and developmental control of CTCF binding site occupancy. We propose that the unique topological configuration of stripe domains represents a permissive landscape facilitating both productive and opportunistic gene regulation and is important for cellular identity.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

