Estimation of Transcription Factor Activity in Knockdown Studies
- PMID: 31270369
- PMCID: PMC6610105
- DOI: 10.1038/s41598-019-46053-7
Estimation of Transcription Factor Activity in Knockdown Studies
Abstract
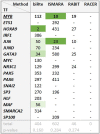
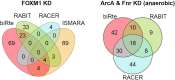
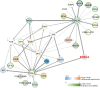
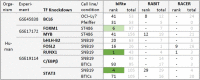
Numerous methods have been developed trying to infer actual regulatory events in a sample. A prominent class of methods model genome-wide gene expression as linear equations derived from a transcription factor (TF) - gene network and optimizes parameters to fit the measured expression intensities. We apply four such methods on experiments with a TF-knockdown (KD) in human and E. coli. The transcriptome data provides clear expression signals and thus represents an extremely favorable test setting. The methods estimate activity changes of all TFs, which we expect to be highest in the KD TF. However, only in 15 out of 54 cases, the KD TFs ranked in the top 5%. We show that this poor overall performance cannot be attributed to a low effectiveness of the knockdown or the specific regulatory network provided as background knowledge. Further, the ranks of regulators related to the KD TF by the network or pathway are not significantly different from a random selection. In general, the result overlaps of different methods are small, indicating that they draw very different conclusions when presented with the same, presumably simple, inference problem. These results show that the investigated methods cannot yield robust TF activity estimates in knockdown schemes.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Reconstructing genome-wide regulatory network of E. coli using transcriptome data and predicted transcription factor activities.BMC Bioinformatics. 2011 Jun 13;12:233. doi: 10.1186/1471-2105-12-233. BMC Bioinformatics. 2011. PMID: 21668997 Free PMC article.
-
The Escherichia coli transcriptome mostly consists of independently regulated modules.Nat Commun. 2019 Dec 4;10(1):5536. doi: 10.1038/s41467-019-13483-w. Nat Commun. 2019. PMID: 31797920 Free PMC article.
-
Diversity of Transcriptional Regulatory Adaptation in E. coli.Mol Biol Evol. 2024 Nov 1;41(11):msae240. doi: 10.1093/molbev/msae240. Mol Biol Evol. 2024. PMID: 39531644 Free PMC article.
-
Building a complete image of genome regulation in the model organism Escherichia coli.J Gen Appl Microbiol. 2018 Jan 15;63(6):311-324. doi: 10.2323/jgam.2017.01.002. Epub 2017 Sep 12. J Gen Appl Microbiol. 2018. PMID: 28904250 Review.
-
New insights into the regulatory networks of paralogous genes in bacteria.Microbiology (Reading). 2010 Jan;156(Pt 1):14-22. doi: 10.1099/mic.0.033266-0. Epub 2009 Oct 22. Microbiology (Reading). 2010. PMID: 19850620 Review.
Cited by
-
A ChIP-exo screen of 887 Protein Capture Reagents Program transcription factor antibodies in human cells.Genome Res. 2021 Sep;31(9):1663-1679. doi: 10.1101/gr.275472.121. Epub 2021 Aug 23. Genome Res. 2021. PMID: 34426512 Free PMC article.
-
Inferring TF activities and activity regulators from gene expression data with constraints from TF perturbation data.Bioinformatics. 2021 Jun 9;37(9):1234-1245. doi: 10.1093/bioinformatics/btaa947. Bioinformatics. 2021. PMID: 33135076 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

