An optimised tissue disaggregation and data processing pipeline for characterising fibroblast phenotypes using single-cell RNA sequencing
- PMID: 31270426
- PMCID: PMC6610623
- DOI: 10.1038/s41598-019-45842-4
An optimised tissue disaggregation and data processing pipeline for characterising fibroblast phenotypes using single-cell RNA sequencing
Abstract
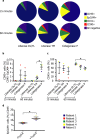
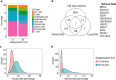
Single-cell RNA sequencing (scRNA-Seq) provides a valuable platform for characterising multicellular ecosystems. Fibroblasts are a heterogeneous cell type involved in many physiological and pathological processes, but remain poorly-characterised. Analysis of fibroblasts is challenging: these cells are difficult to isolate from tissues, and are therefore commonly under-represented in scRNA-seq datasets. Here, we describe an optimised approach for fibroblast isolation from human lung tissues. We demonstrate the potential for this procedure in characterising stromal cell phenotypes using scRNA-Seq, analyse the effect of tissue disaggregation on gene expression, and optimise data processing to improve clustering quality. We also assess the impact of in vitro culture conditions on stromal cell gene expression and proliferation, showing that altering these conditions can skew phenotypes.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Zepp Jarod A., Zacharias William J., Frank David B., Cavanaugh Christina A., Zhou Su, Morley Michael P., Morrisey Edward E. Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell. 2017;170(6):1134-1148.e10. doi: 10.1016/j.cell.2017.07.034. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

