A CC-NBS-LRR gene induces hybrid lethality in cotton
- PMID: 31270546
- PMCID: PMC6793457
- DOI: 10.1093/jxb/erz312
A CC-NBS-LRR gene induces hybrid lethality in cotton
Abstract
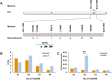
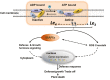
Hybrid lethality forms a reproductive barrier that has been found in many eukaryotes. Most cases follow the Bateson-Dobzhansky-Muller genetic incompatibility model and involve two or more loci. In this study, we demonstrate that a coiled-coil nucleotide-binding site leucine-rich repeat (CC-NBS-LRR) gene is the causal gene underlying the Le4 locus for interspecific hybrid lethality between Gossypium barbadense and G. hirsutum (cotton). Silencing this CC-NBS-LRR gene can restore F1 plants from a lethal to a normal phenotype. A total of 11 099 genes were differentially expressed between the leaves of normal and lethal F1 plants, of which genes related to autoimmune responses were highly enriched. Genes related to ATP-binding and ATPase were up-regulated before the lethal syndrome appeared; this may result in the conversion of Le4 into an active state and hence trigger immune signals in the absence of biotic/abiotic stress. We discuss our results in relation to the evolution and domestication of Sea Island cottons and the molecular mechanisms of hybrid lethality associated with autoimmune responses. Our findings provide new insights into reproductive isolation and may benefit cotton breeding.
Keywords: ATP-binding; DNA-dependent ATPase activity; NBS-LRR genes; autoimmunity response; cotton; hybrid lethality; map-based cloning.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



Similar articles
-
A recessive LRR-RLK gene causes hybrid breakdown in cotton.Theor Appl Genet. 2023 Aug 15;136(9):189. doi: 10.1007/s00122-023-04427-6. Theor Appl Genet. 2023. PMID: 37582982
-
Comparative analysis of resistance gene analogues encoding NBS-LRR domains in cotton.J Sci Food Agric. 2016 Jan 30;96(2):530-8. doi: 10.1002/jsfa.7120. Epub 2015 Mar 3. J Sci Food Agric. 2016. PMID: 25640313
-
Interaction of novel Dobzhansky-Muller type genes for the induction of hybrid lethality between Gossypium hirsutum and G. barbadense cv. Coastland R4-4.Theor Appl Genet. 2009 Jun;119(1):33-41. doi: 10.1007/s00122-009-1014-5. Epub 2009 Mar 28. Theor Appl Genet. 2009. PMID: 19330312
-
Understanding and overcoming hybrid lethality in seed and seedling stages as barriers to hybridization and gene flow.Front Plant Sci. 2023 Jul 5;14:1219417. doi: 10.3389/fpls.2023.1219417. eCollection 2023. Front Plant Sci. 2023. PMID: 37476165 Free PMC article. Review.
-
One Hundred Years of Hybrid Necrosis: Hybrid Autoimmunity as a Window into the Mechanisms and Evolution of Plant-Pathogen Interactions.Annu Rev Phytopathol. 2021 Aug 25;59:213-237. doi: 10.1146/annurev-phyto-020620-114826. Epub 2021 May 4. Annu Rev Phytopathol. 2021. PMID: 33945695 Review.
Cited by
-
Two NLR immune receptors acquired high-affinity binding to a fungal effector through convergent evolution of their integrated domain.Elife. 2021 Jul 21;10:e66961. doi: 10.7554/eLife.66961. Elife. 2021. PMID: 34288868 Free PMC article.
-
Genetic Mapping of the HLA1 Locus Causing Hybrid Lethality in Nicotiana Interspecific Hybrids.Plants (Basel). 2021 Sep 30;10(10):2062. doi: 10.3390/plants10102062. Plants (Basel). 2021. PMID: 34685871 Free PMC article.
-
Analysis of the possible cytogenetic mechanism for overcoming hybrid lethality in an interspecific cross between Nicotiana suaveolens and Nicotiana tabacum.Sci Rep. 2021 Apr 9;11(1):7812. doi: 10.1038/s41598-021-87242-7. Sci Rep. 2021. PMID: 33837225 Free PMC article.
-
Transcriptome and plant hormone analyses provide new insight into the molecular regulatory networks underlying hybrid lethality in cabbage (Brassica oleracea).Planta. 2021 Apr 11;253(5):96. doi: 10.1007/s00425-021-03608-1. Planta. 2021. PMID: 33839925
-
Genome-Wide Identification and Characterization of the CC-NBS-LRR Gene Family in Cucumber (Cucumis sativus L.).Int J Mol Sci. 2022 May 2;23(9):5048. doi: 10.3390/ijms23095048. Int J Mol Sci. 2022. PMID: 35563438 Free PMC article.
References
-
- Alcázar R, García AV, Kronholm I, de Meaux J, Koornneef M, Parker JE, Reymond M. 2010. Natural variation at Strubbelig Receptor Kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nature Genetics 42, 1135–1139. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

