Functional analysis of LHCSR1, a protein catalyzing NPQ in mosses, by heterologous expression in Arabidopsis thaliana
- PMID: 31270669
- PMCID: PMC6874524
- DOI: 10.1007/s11120-019-00656-3
Functional analysis of LHCSR1, a protein catalyzing NPQ in mosses, by heterologous expression in Arabidopsis thaliana
Abstract
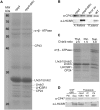
Non-photochemical quenching, NPQ, of chlorophyll fluorescence regulates the heat dissipation of chlorophyll excited states and determines the efficiency of the oxygenic photosynthetic systems. NPQ is regulated by a pH-sensing protein, responding to the chloroplast lumen acidification induced by excess light, coupled to an actuator, a chlorophyll/xanthophyll subunit where quenching reactions are catalyzed. In plants, the sensor is PSBS, while the two pigment-binding proteins Lhcb4 (also known as CP29) and LHCII are the actuators. In algae and mosses, stress-related light-harvesting proteins (LHCSR) comprise both functions of sensor and actuator within a single subunit. Here, we report on expressing the lhcsr1 gene from the moss Physcomitrella patens into several Arabidopsis thaliana npq4 mutants lacking the pH sensing PSBS protein essential for NPQ activity. The heterologous protein LHCSR1 accumulates in thylakoids of A. thaliana and NPQ activity can be partially restored. Complementation of double mutants lacking, besides PSBS, specific xanthophylls, allowed analyzing chromophore requirement for LHCSR-dependent quenching activity. We show that the partial recovery of NPQ is mostly due to the lower levels of Zeaxanthin in A. thaliana in comparison to P. patens. Complemented npq2npq4 mutants, lacking besides PSBS, Zeaxanthin Epoxidase, showed an NPQ recovery of up to 70% in comparison to A. thaliana wild type. Furthermore, we show that Lutein is not essential for the folding nor for the quenching activity of LHCSR1. In short, we have developed a system to study the function of LHCSR proteins using heterologous expression in a variety of A. thaliana mutants.
Keywords: A. thaliana; Heterologous expression; LHCSR; NPQ; P. patens; Photoprotection.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






Similar articles
-
Heterologous expression of moss light-harvesting complex stress-related 1 (LHCSR1), the chlorophyll a-xanthophyll pigment-protein complex catalyzing non-photochemical quenching, in Nicotiana sp.J Biol Chem. 2015 Oct 2;290(40):24340-54. doi: 10.1074/jbc.M115.668798. Epub 2015 Aug 10. J Biol Chem. 2015. PMID: 26260788 Free PMC article.
-
Coexistence of plant and algal energy dissipation mechanisms in the moss Physcomitrella patens.New Phytol. 2012 Nov;196(3):763-773. doi: 10.1111/j.1469-8137.2012.04345.x. Epub 2012 Sep 25. New Phytol. 2012. PMID: 23005032
-
The rise and fall of Light-Harvesting Complex Stress-Related proteins as photoprotection agents during evolution.J Exp Bot. 2019 Oct 24;70(20):5527-5535. doi: 10.1093/jxb/erz317. J Exp Bot. 2019. PMID: 31424076
-
Molecular genetics of xanthophyll-dependent photoprotection in green algae and plants.Philos Trans R Soc Lond B Biol Sci. 2000 Oct 29;355(1402):1385-94. doi: 10.1098/rstb.2000.0700. Philos Trans R Soc Lond B Biol Sci. 2000. PMID: 11127993 Free PMC article. Review.
-
Energy-Dependent Non-Photochemical Quenching: PsbS, LhcSR, and Other Players.Biochemistry (Mosc). 2025 Jan;90(1):44-60. doi: 10.1134/S000629792460371X. Biochemistry (Mosc). 2025. PMID: 40058973 Review.
Cited by
-
Long-term adaptation of Arabidopsis thaliana to far-red light.Plant Cell Environ. 2021 Sep;44(9):3002-3014. doi: 10.1111/pce.14032. Epub 2021 May 5. Plant Cell Environ. 2021. PMID: 33599977 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

