A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation
- PMID: 31273116
- PMCID: PMC7062386
- DOI: 10.1126/science.aau9263
A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation
Abstract
The COPII-cargo adaptor complex Lst1-Sec23 selectively sorts proteins into vesicles that bud from the endoplasmic reticulum (ER) and traffic to the Golgi. Improperly folded proteins are prevented from exiting the ER and are degraded. ER-phagy is an autophagic degradation pathway that uses ER-resident receptors. Working in yeast, we found an unexpected role for Lst1-Sec23 in ER-phagy that was independent from its function in secretion. Up-regulation of the stress-inducible ER-phagy receptor Atg40 induced the association of Lst1-Sec23 with Atg40 at distinct ER domains to package ER into autophagosomes. Lst1-mediated ER-phagy played a vital role in maintaining cellular homeostasis by preventing the accumulation of an aggregation-prone protein in the ER. Lst1 function appears to be conserved because its mammalian homolog, SEC24C, was also required for ER-phagy.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
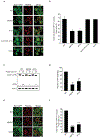
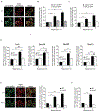
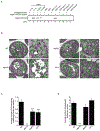


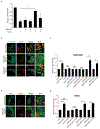
References
-
- Kirchhausen T, Bonifacino JS, Riezman H, Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr Opin Cell Biol 9, 488–495 (1997) - PubMed
-
- Tang BL, Kausalya J, Low DY, Lock ML, Hong W, A family of mammalian proteins homologous to yeast Sec24p. Biochem Biophys Res Commun 258, 679–684 (1999). - PubMed
-
- Walter P, Ron D, The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

