Human Corneal Expression of SLC4A11, a Gene Mutated in Endothelial Corneal Dystrophies
- PMID: 31273259
- PMCID: PMC6609610
- DOI: 10.1038/s41598-019-46094-y
Human Corneal Expression of SLC4A11, a Gene Mutated in Endothelial Corneal Dystrophies
Abstract
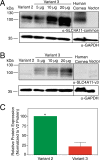
Two blinding corneal dystrophies, pediatric-onset congenital hereditary endothelial dystrophy (CHED) and some cases of late-onset Fuchs endothelial corneal dystrophy (FECD), are caused by SLC4A11 mutations. Three N-terminal SLC4A11 variants: v1, v2 and v3 are expressed in humans. We set out to determine which of these transcripts and what translated products, are present in corneal endothelium as these would be most relevant for CHED and FECD studies. Reverse transcription PCR (RT-PCR) and quantitative RT-PCR revealed only v2 and v3 mRNA in human cornea, but v2 was most abundant. Immunoblots probed with variant-specific antibodies revealed that v2 protein is about four times more abundant than v3 in human corneal endothelium. Bioinformatics and protein analysis using variant-specific antibodies revealed that second methionine in the open reading frame (M36) acts as translation initiation site on SLC4A11 v2 in human cornea. The v2 variants starting at M1 (v2-M1) and M36 (v2-M36) were indistinguishable in their cell surface trafficking and transport function (water flux). Structural homology models of v2-M36 and v3 suggest structural differences but their significance remains unclear. A combination of bioinformatics, RNA quantification and isoform-specific antibodies allows us to conclude that SLC4A11 variant 2 with start site M36 is predominant in corneal endothelium.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Antioxidant MitoQ increases viability of human corneal endothelial cells with congenital hereditary endothelial dystrophy-associated SLC4A11 mutations.Ophthalmic Genet. 2025 Apr;46(2):166-173. doi: 10.1080/13816810.2025.2450455. Epub 2025 Jan 20. Ophthalmic Genet. 2025. PMID: 39834031
-
SLC4A11 Three-Dimensional Homology Model Rationalizes Corneal Dystrophy-Causing Mutations.Hum Mutat. 2017 Mar;38(3):279-288. doi: 10.1002/humu.23152. Epub 2016 Dec 27. Hum Mutat. 2017. PMID: 27925686
-
Ophthalmic Nonsteroidal Anti-Inflammatory Drugs as a Therapy for Corneal Dystrophies Caused by SLC4A11 Mutation.Invest Ophthalmol Vis Sci. 2018 Aug 1;59(10):4258-4267. doi: 10.1167/iovs.18-24301. Invest Ophthalmol Vis Sci. 2018. PMID: 30140924
-
SLC4A11 and the Pathophysiology of Congenital Hereditary Endothelial Dystrophy.Biomed Res Int. 2015;2015:475392. doi: 10.1155/2015/475392. Epub 2015 Sep 16. Biomed Res Int. 2015. PMID: 26451371 Free PMC article. Review.
-
Update on the genetics of corneal endothelial dystrophies.Indian J Ophthalmol. 2022 Jul;70(7):2239-2248. doi: 10.4103/ijo.IJO_992_22. Indian J Ophthalmol. 2022. PMID: 35791103 Free PMC article. Review.
Cited by
-
Animal models of corneal endothelial dysfunction to facilitate development of novel therapies.Ann Transl Med. 2021 Aug;9(15):1271. doi: 10.21037/atm-20-4389. Ann Transl Med. 2021. PMID: 34532408 Free PMC article. Review.
-
Investigation of the functional impact of CHED- and FECD4-associated SLC4A11 mutations in human corneal endothelial cells.PLoS One. 2024 Jan 22;19(1):e0296928. doi: 10.1371/journal.pone.0296928. eCollection 2024. PLoS One. 2024. PMID: 38252645 Free PMC article.
-
Energy Shortage in Human and Mouse Models of SLC4A11-Associated Corneal Endothelial Dystrophies.Invest Ophthalmol Vis Sci. 2020 Jul 1;61(8):39. doi: 10.1167/iovs.61.8.39. Invest Ophthalmol Vis Sci. 2020. PMID: 32721020 Free PMC article.
-
Genetic mutations and molecular mechanisms of Fuchs endothelial corneal dystrophy.Eye Vis (Lond). 2021 Jun 15;8(1):24. doi: 10.1186/s40662-021-00246-2. Eye Vis (Lond). 2021. PMID: 34130750 Free PMC article. Review.
-
Diseases of the corneal endothelium.Exp Eye Res. 2021 Apr;205:108495. doi: 10.1016/j.exer.2021.108495. Epub 2021 Feb 14. Exp Eye Res. 2021. PMID: 33596440 Free PMC article. Review.
References
-
- 2016 Eye Banking Statisitical Report. (Eye Bank Association of America, Washington, D.C., U.S.A., 2017).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

