Cas9-based enrichment and single-molecule sequencing for precise characterization of genomic duplications
- PMID: 31273287
- PMCID: PMC6923135
- DOI: 10.1038/s41374-019-0283-0
Cas9-based enrichment and single-molecule sequencing for precise characterization of genomic duplications
Abstract
The widespread use of genome-wide diagnostic screening methods has greatly increased the frequency with which incidental (but possibly pathogenic) copy number changes affecting single genes are detected. These findings require validation to allow appropriate clinical management. Deletion variants can usually be readily validated using a range of short-read next-generation sequencing (NGS) strategies, but the characterization of duplication variants at nucleotide resolution remains challenging. This presents diagnostic problems, since pathogenicity cannot generally be assessed without knowing the structure of the variant. We have used a novel Cas9 enrichment strategy, in combination with long-read single-molecule nanopore sequencing, to address this need. We describe the nucleotide-level resolution of two problematic cases, both of whom presented with neurodevelopmental problems and were initially investigated by array CGH. In the first case, an incidental 1.7-kb imbalance involving a partial duplication of VHL exon 3 was detected. This variant was inherited from the patient's father, who had a history of renal cancer at 38 years. In the second case, an incidental ~200-kb de novo duplication that included DMD exons 30-44 was resolved. In both cases, the long-read data yielded sufficient information to enable Sanger sequencing to define the rearrangement breakpoints, and creation of breakpoint-spanning PCR assays suitable for testing of relatives. Our Cas9 enrichment and nanopore sequencing approach can be readily adopted by molecular diagnostic laboratories for cost-effective and rapid characterization of challenging duplication-containing alleles. We also anticipate that in future this method may prove useful for characterizing acquired translocations in tumor cells, and for precisely identifying transgene integration sites in mouse models.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
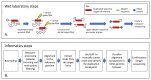
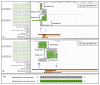
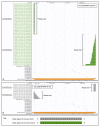

References
-
- Davies SC. Annual Report of the Chief Medical Officer 2016, Generation Genome. London: Department of Health; 2017. [cited 7 February 2019]. [Internet], Available from https://assets.publishing.service.gov.uk/government/uploads/system/uploa....
-
- van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The Third Revolution in Sequencing Technology. Trends Genet. 2018;34:666–681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

